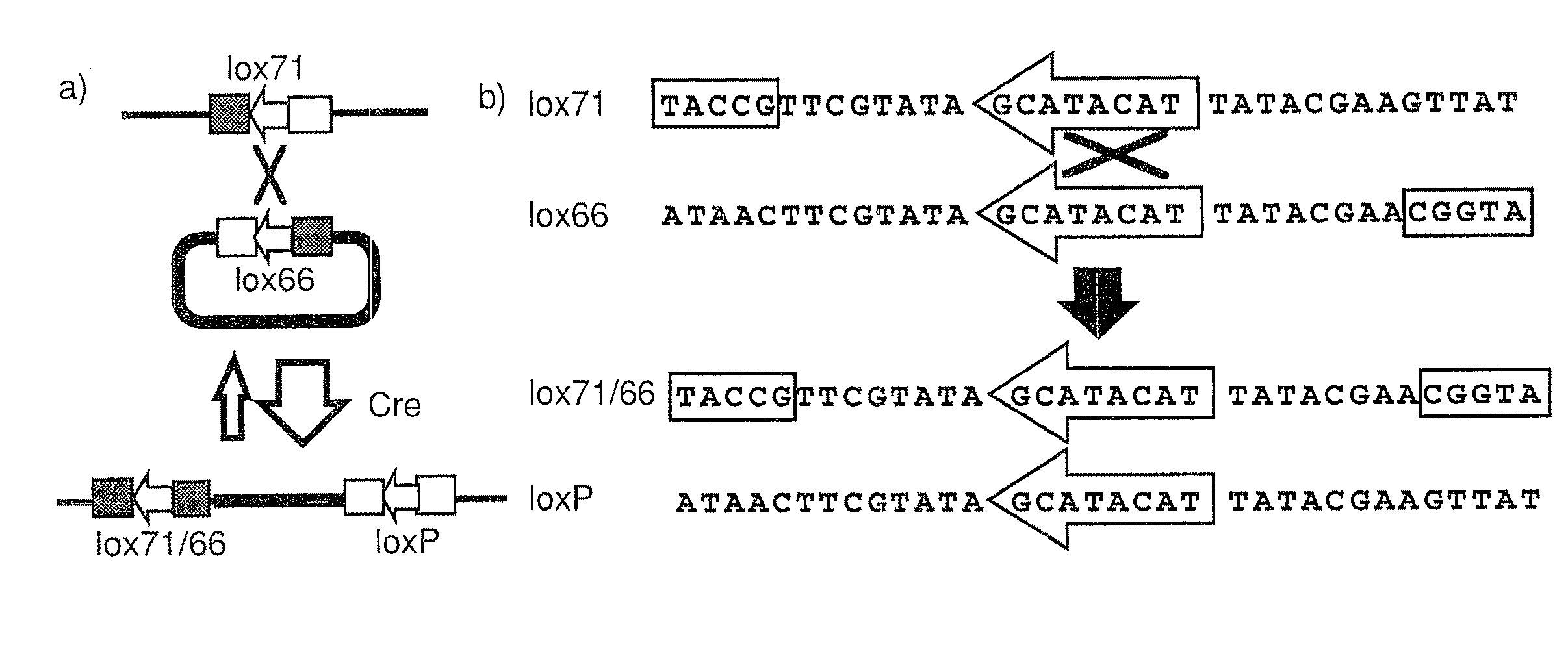
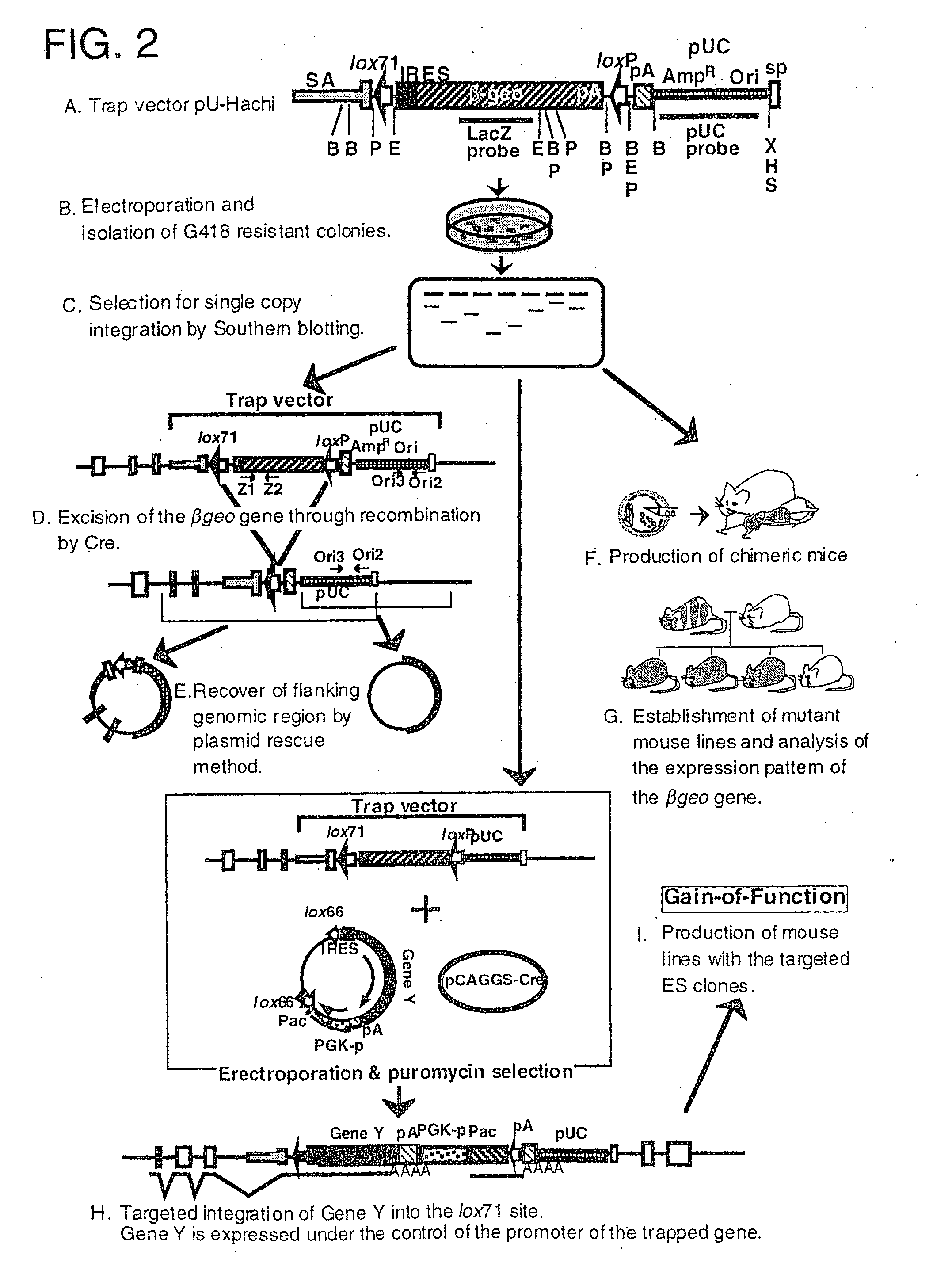
Trap vectors and gene trapping using the same
a gene and vector technology, applied in the field of random mutation es clone technology using gene trapping, can solve the problems of insufficient information on functions, inability to explain diseases with the structure of causative genes alone, and inability to analyze the functions of elements to which the transcription factor binds at present, so as to achieve easy handling and propagation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction of Varied-Type Gene Trap Vectors
[0126](1) Construction of pU-Hachi Trap Vector
[0127]pU-Hachi vector is derived from pGT1.8IRES β-geo, and contains SA sequence from mouse En-2 gene and β-geo sequence linked to encephalomyocarditis virus-derived IRES sequence. First, a BamHI fragment of lox71 is inserted into the BglII site of pGT1.8RES β-geo. Then, plasmid pEBN-SE7ti was constructed by inserting a 180 bp (SP) sequence (which is a part of rabbit β globin gene), loxP sequence, and poly A addition signal from mouse phosphoglycerate kinase-1 (PGK) into a modified vector pUC19 from which lacZ sequence has been removed. The SP sequence was used to protect the 3′ end of the trap vector. By inserting a SalI fragment of SA-IRES-lox71-β-geo into the SalI site of pEBN-SE7ti, pU-Hachi was obtained.
(2) Construction of pU-12 Trap Vector
[0128]In order to construct pU-12 trap vector, first, the PGK poly(A) signal of pE3NSE7 was replaced with puromycin resistance gene+PGK poly(A) signal....
example 2
Selection of ES Cell Clones
[0130]In the electroporation using pU-Hachi trap vector, 100 μg of SpeI-digested DNA and 3×107 cells were used. Cells were suspended in 0.8 ml of PBS and electroporated using a BioRad GenePulser at 800 V and 3μF. After 48 hours, the cells were cultured in the presence of 200 μg / ml G418. This selection was maintained for 7 days. The resultant colonies were plated on 24-well plates for propagation and stored frozen. Trap clones were analyzed by Southern blotting to select those cell strains that exhibit patterns of single copy integration.
[0131]In order to remove β-geo sequence from the trap clone, pCAGGS-Cre (Araki, K. et al., Proc. Natl. Acad. Sci. USA, 92:160-164, 1995; Araki, K. et al., Nucl. Acids Res., 25:868-872, 1997; Araki, K. et al., J. Biochem. Tokyo, 122: 977-982, 1997) was electroporated in a circular form. This electroporation was carried out under the same conditions as described above except that the number of cells was 1.5×107 and that the P...
example 3
Selection Frequency of Clones
[0140]In order to select those clones in which a single copy of the trap vector was integrated, DNA was extracted from the selected, neomycin resistant clones and analyzed by Southern blotting.
[0141]Briefly, cells were lysed with SDS / proteinase K, treated with phenol / chloroform (1:1, vol:vol) twice, precipitated with ethanol, and then dissolved in TE buffer (10 mM Tris-HCl, pH 7.5 / 1 mM EDTA). Six micrograms of genomic DNA was digested with appropriate restriction enzymes, electrophoresed on 0.9% agarose gel and then blotted onto a nylon membrane (Boehringer Mannheim). Hybridization was performed using a DIG DNA Labeling and Detection Kit (Boehringer Mannheim).
[0142]For PCR analysis, DNA was subjected to 28 cycles of denaturation at 94° C. for 1 min, annealing at 55° C. for 2 min and extension at 72° C. for 2 min in the reaction solution described below.
[0143]The primers used in the PCR were as follows:
β-geo detection primers
Z1 (forward):5′-gcgttacccaactt...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com