Stabilisation of biological samples
a biological sample and stabilisation technology, applied in the field of biological sample stabilisation, can solve the problems of severe alteration of the expression profile of the targeted molecules, compromise of subsequent analysis, and inability to achieve any practicable methods in hospitals, doctor surgeries or diagnostic routine laboratories, so as to reduce the yield of such nucleic acids and reduce the recovery of such stabilized nucleic acids
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
tion Using Formamide
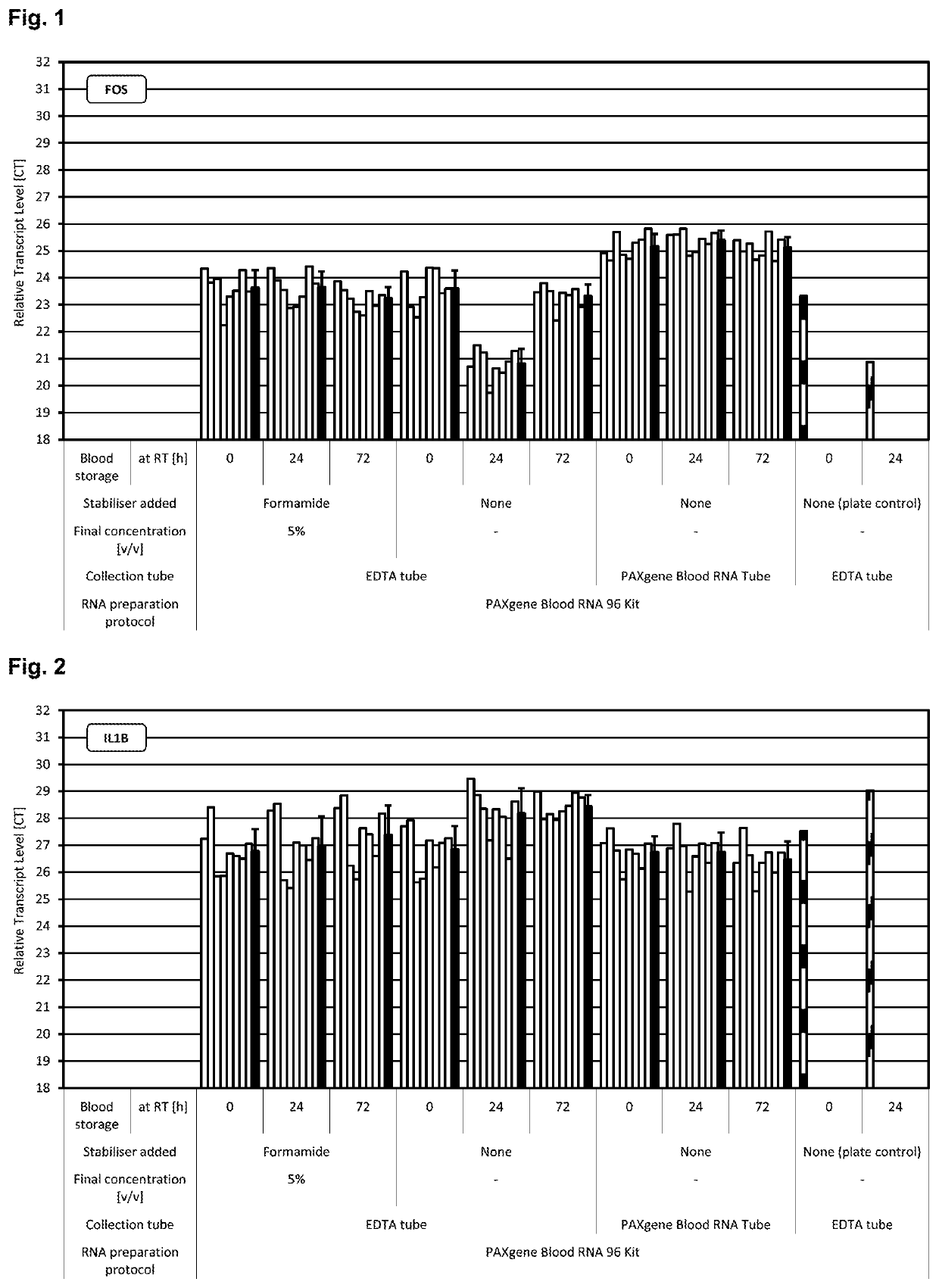
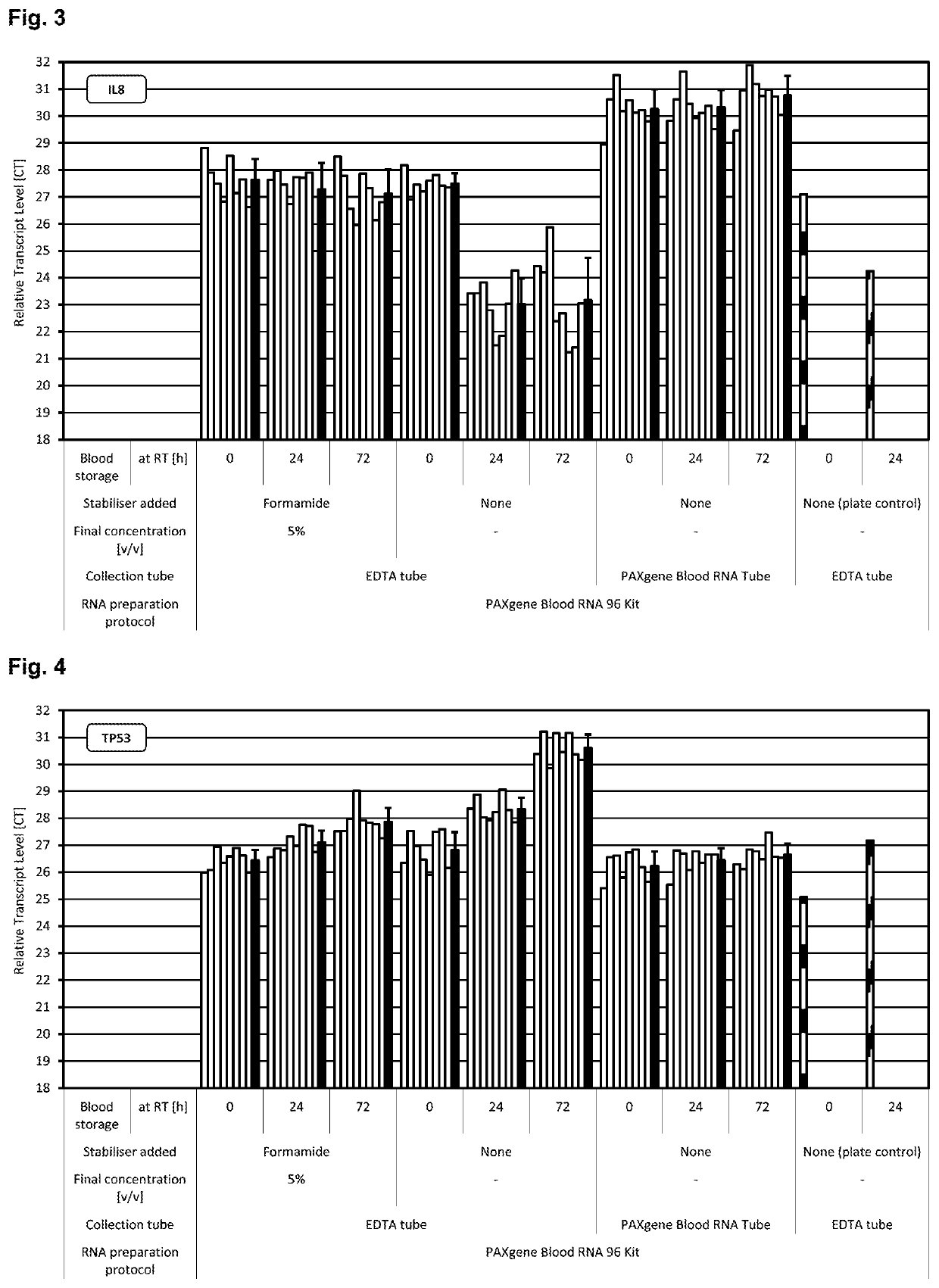
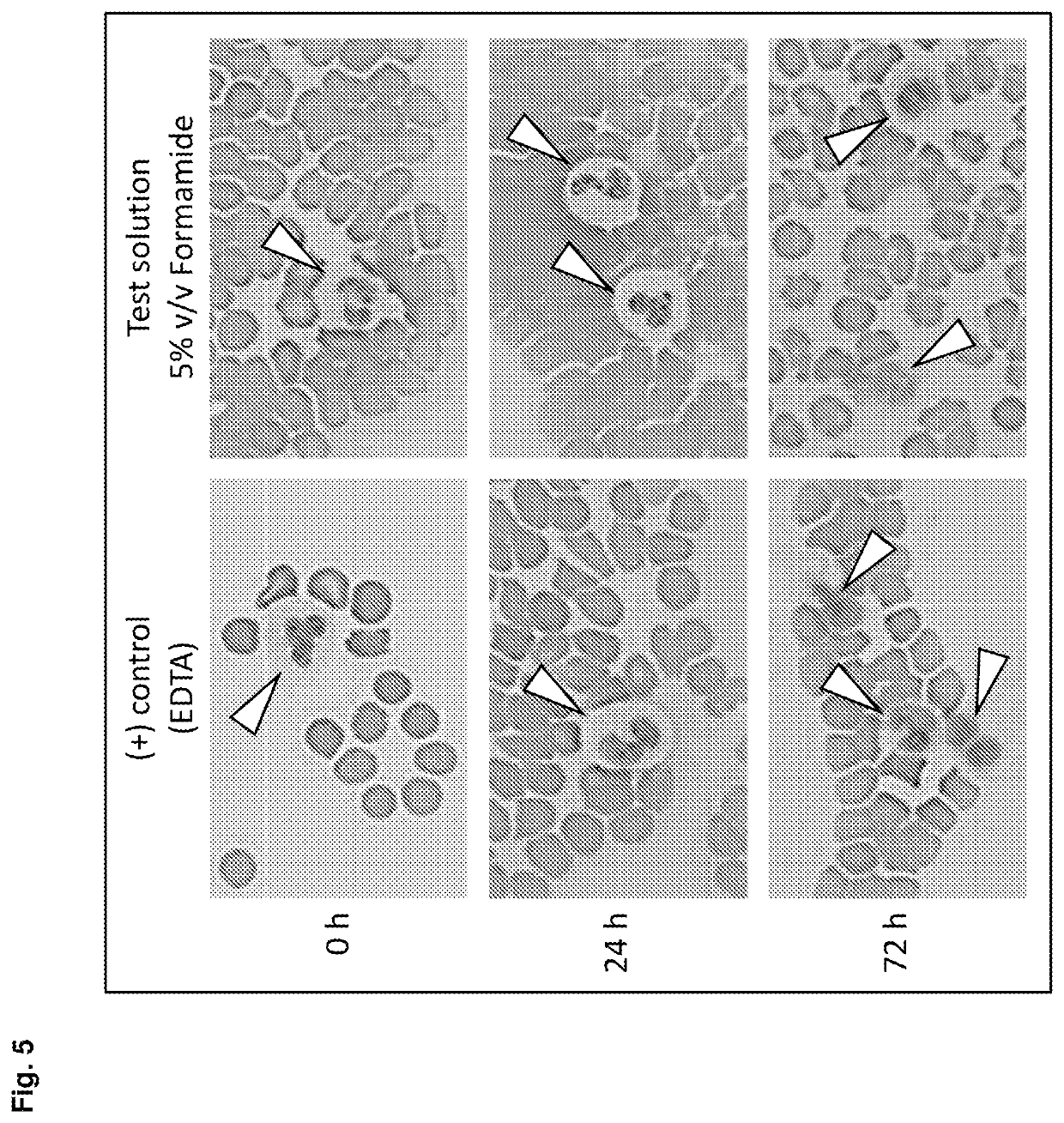
[0280]Blood was collected from multiple donors into replicate EDTA blood tubes (BD, [−] control of sample stabilisation) and PAXgene Blood RNA Tubes (PreAnalytiX) serving as [+] control of sample stabilisation. Immediately after blood collection half of the EDTA blood samples were treated with RNA stabilisation test solution, resulting in blood test samples. In detail, 1 ml stabilisation additive (50% v / v formamide, 5×MOPS buffer, pH5.5) was added to 9 ml EDTA blood. Blood mixing, incubation, transfer of blood sample aliquots to PAXgene Blood RNA Tubes, freezing, storage and RNA preparation was done as described above. RNA was subjected to transcript level analysis as described above in Material and methods. Quantity (RNA yield) and quality (RNA purity, RNA integrity) of RNA was measured by UV spectroscopy and miniaturised capillary gel electrophoresis with RIN calculation (Nanochips on Agilent Bioanalyzer). Transcript levels of all RNA samples were analysed by r...
example 2
tion Using Acetamide
[0286]EDTA blood samples were kept untreated or mixed with 8% w / v acetamide immediately after blood collection. Blood samples collected in PAXgene RNA blood tubes served again as control. Sample handling and RNA isolation was performed from blood samples without and from replicate tubes after incubation for one day at RT as described in material and methods. Transcript levels of all RNA samples were analysed as described in material and methods by real time RT-PCR using monoplex assays of FOS, IL1B, IL8 and TP53, normalized to the amount of template input into the reaction. The results are shown in FIGS. 7 to 10. Resulting CT values reflecting the amount of transcripts were directly compared. Relative transcripts levels unaffected from blood sample incubation at RT were indicated by constant CT values, while gains of transcripts (e.g., by gene induction) were indicated by lower CT and losses of transcripts (e.g., by gene repression) by higher CT values. As can be...
example 3
tion Using N-Methylacetamide
[0287]Samples were prepared and processed as described in example 2, however using 2% (w / v) N-methylacetamide as stabilizer in the composition comprising the blood sample and the stabilising agent. The results are shown in FIGS. 11 to 14. As can be seen, the secondary carboxylic acid amide N-methylacetamide was effective in stabilizing transcript levels for up to three days.
[0288]Examples 1 to 3 show that different primary and secondary carboxylic acid amides are highly effective in stabilizing the gene transcription profile of cells in blood samples.
[0289]II. Stabilization of the Extracellular Nucleic Acid Population in Blood Samples Using Different Primary and Secondary Carboxylic Acid Amides
[0290]Materials and Methods
[0291]Different primary and secondary carboxylic acid amides were tested for their ability to stabilize a cell-containing biological sample, here a whole blood sample, either alone or in combination with a caspase inhibitor. As can be seen...
PUM
| Property | Measurement | Unit |
|---|---|---|
| time | aaaaa | aaaaa |
| real time monoplex RT- | aaaaa | aaaaa |
| real time monoplex RT- | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com