Alu sequence of Africa green monkey for detecting content of residual DNA in SARS vaccine
An alu sequence and vaccine technology, applied in the field of Alu sequence of African green monkeys, can solve problems such as undeveloped probes, and achieve the effect of monitoring residual DNA content
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1 Preparation of African Green Monkey Alu Sequence
[0022] PCR method to amplify Alu sequence
[0023] Using the African green monkey Alu sequence X01476 in GeneBank as a reference design PCR primers. Upstream primer P1: 5'-gct, ttg, agg, ccg, ggc, ggg, atg-3' (21bp, SEQ ID NO: 3), downstream primer P2: 5'-ctc, tct, ctt, aga, gtc, ttg , Ctc, tg-3' (23bp, SEQID NO: 4). The PCR reaction parameters are: denaturation at 98°C for 5 minutes, 25sec at 94°C, 30sec at 68°C, 30sec at 72°C, 30 cycles, and extension at 72°C for 10 minutes. After PCR amplification, a 290bp band ( figure 1 ).
[0024] (2) Cloning of Alu sequence
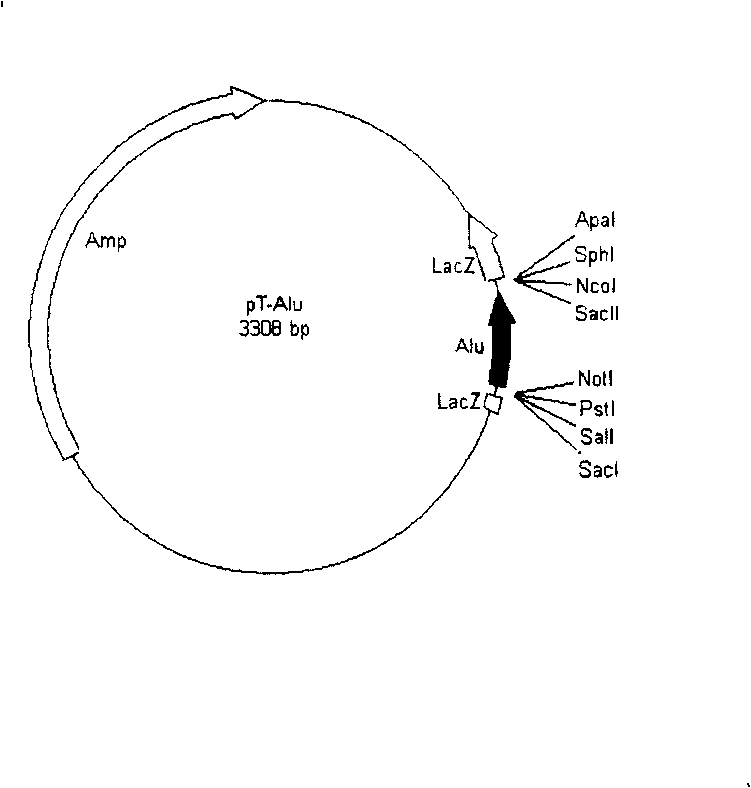
[0025] The PCR product was cloned on Promega's pGEM-T and transformed into E.coli JM109 strain. After a small amount of plasmid was extracted, NcoI and SalI were identified by double enzyme digestion, and several positive clones containing 290bp insert were obtained ( figure 2 ), select a clone named T-Alu, its structure is as image 3 Shown.
Embodiment 2
[0026] Example 2 Method for detecting residual DNA in vaccine by using African green monkey Alu sequence
[0027] 300ng purified and recovered Alu sequence NcoI and SalI digested fragments were labeled according to the instructions of Roche's Digoxin DNA Labeling and Detection Kit. The detection of residual DNA in SARS vaccine adopts dot blot method.
[0028] First, cut a piece of nylon membrane of the required size, respectively spot the Ver0DNA reference standards of 10gn, 1ng, 100pg, and 10pg on the membrane, and then spot the sample to be tested on the membrane according to different dilutions. The membranes were respectively subjected to denaturation solution (0.5M NaOH, 1.5M NaCl), neutralization solution (0.5M Tris pH7.0, 1M NaCl), 2XSSC for 5 minutes each, and then crosslinked in a UV crosslinker at 45J for 5 minutes. The membrane was pre-hybridized in the hybridization bag for 1 hour, and then probes were added for hybridization overnight. After washing the membrane, the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com