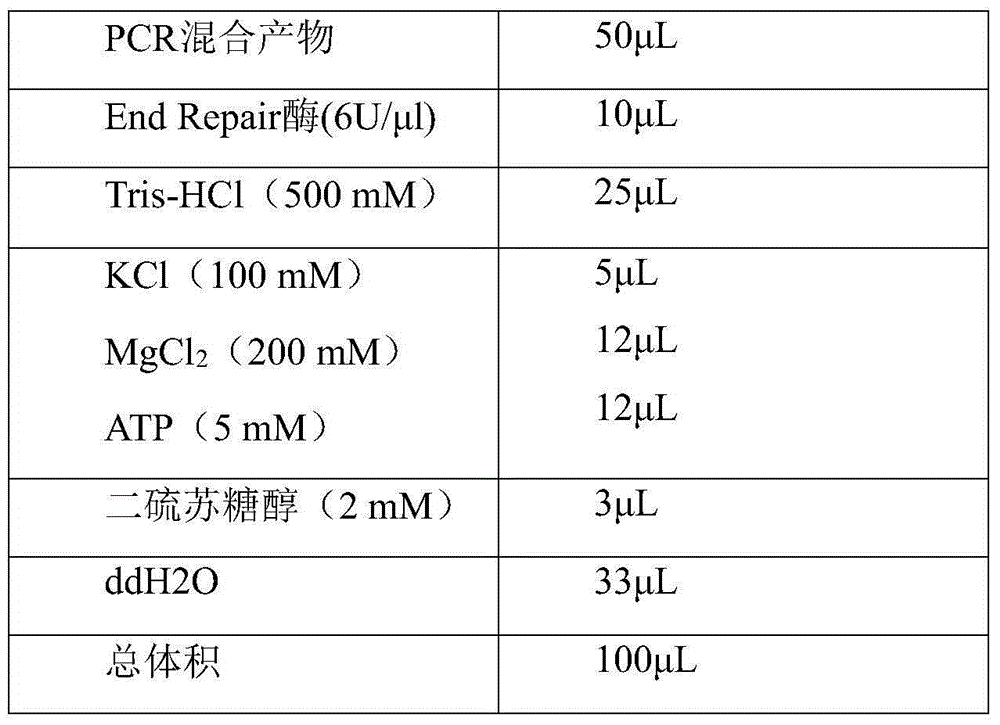
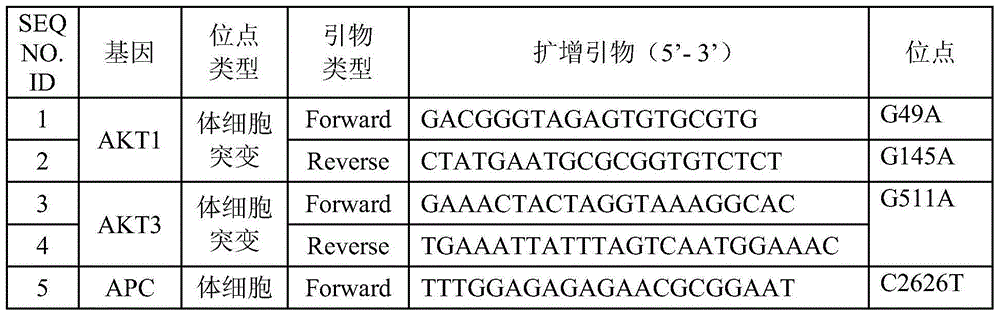
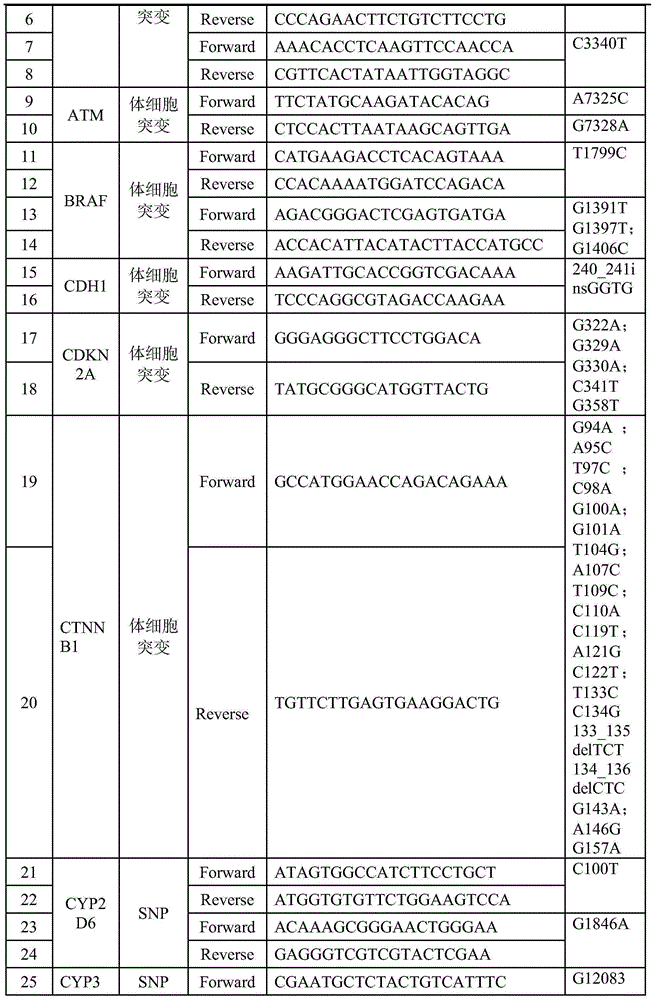
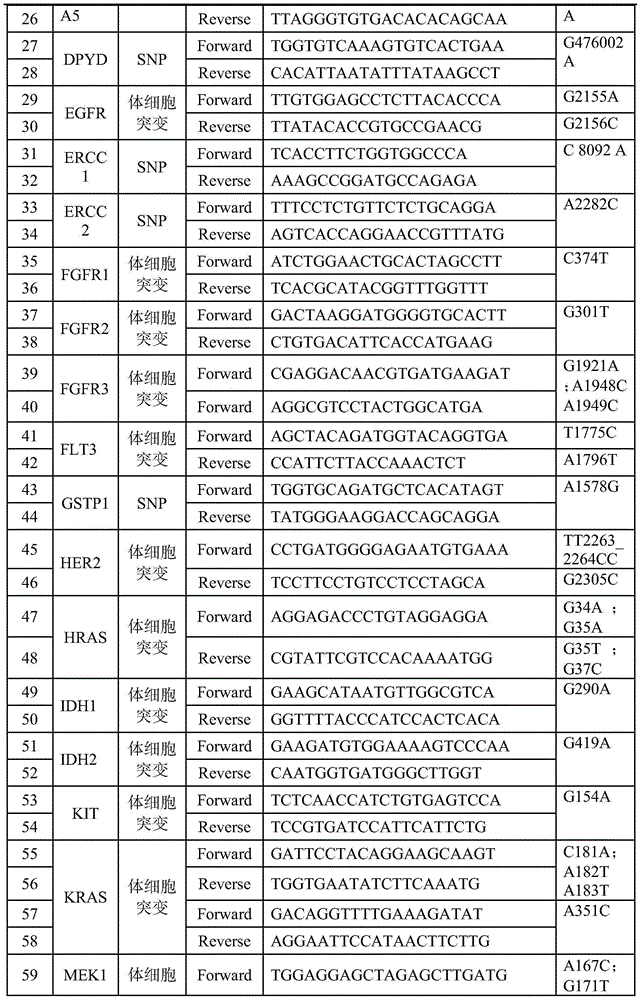
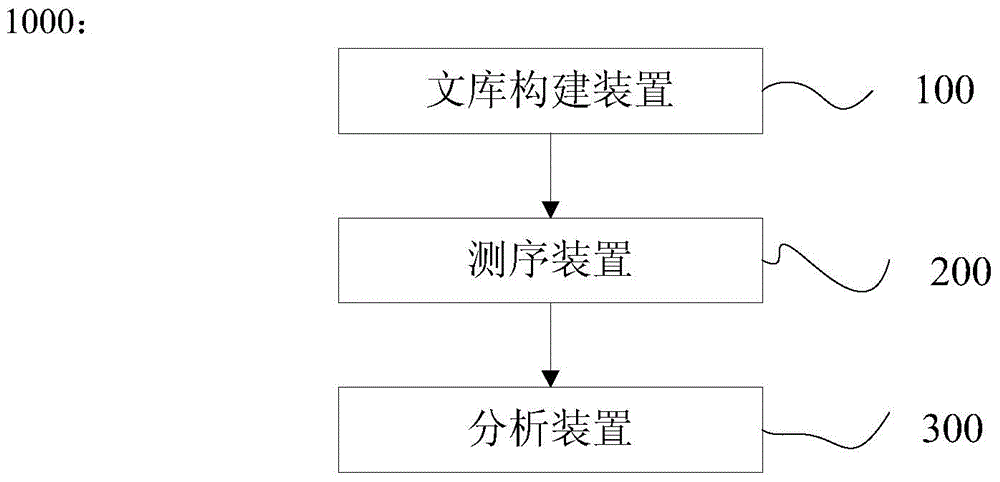
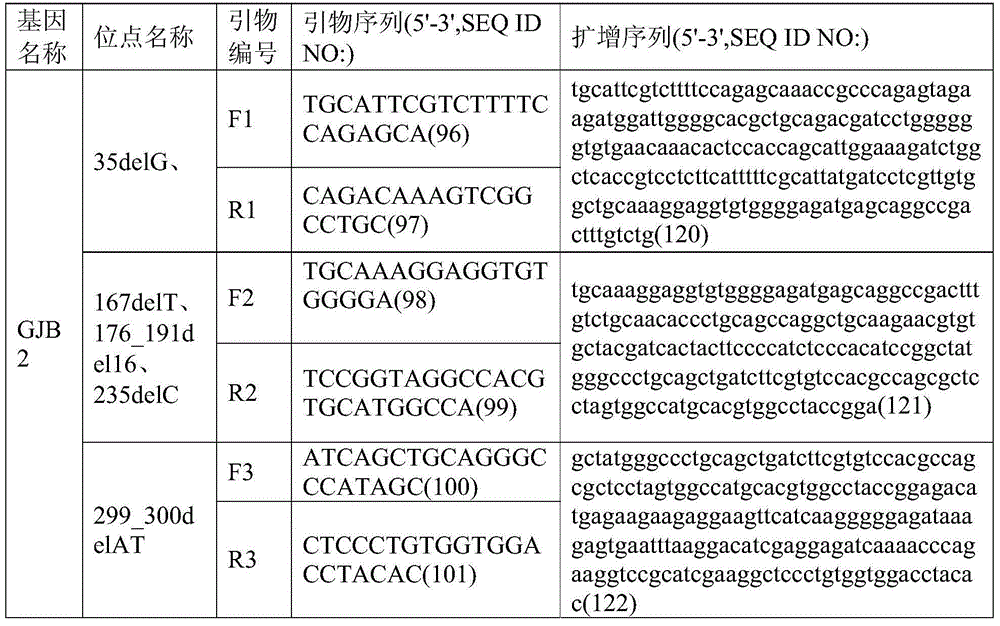
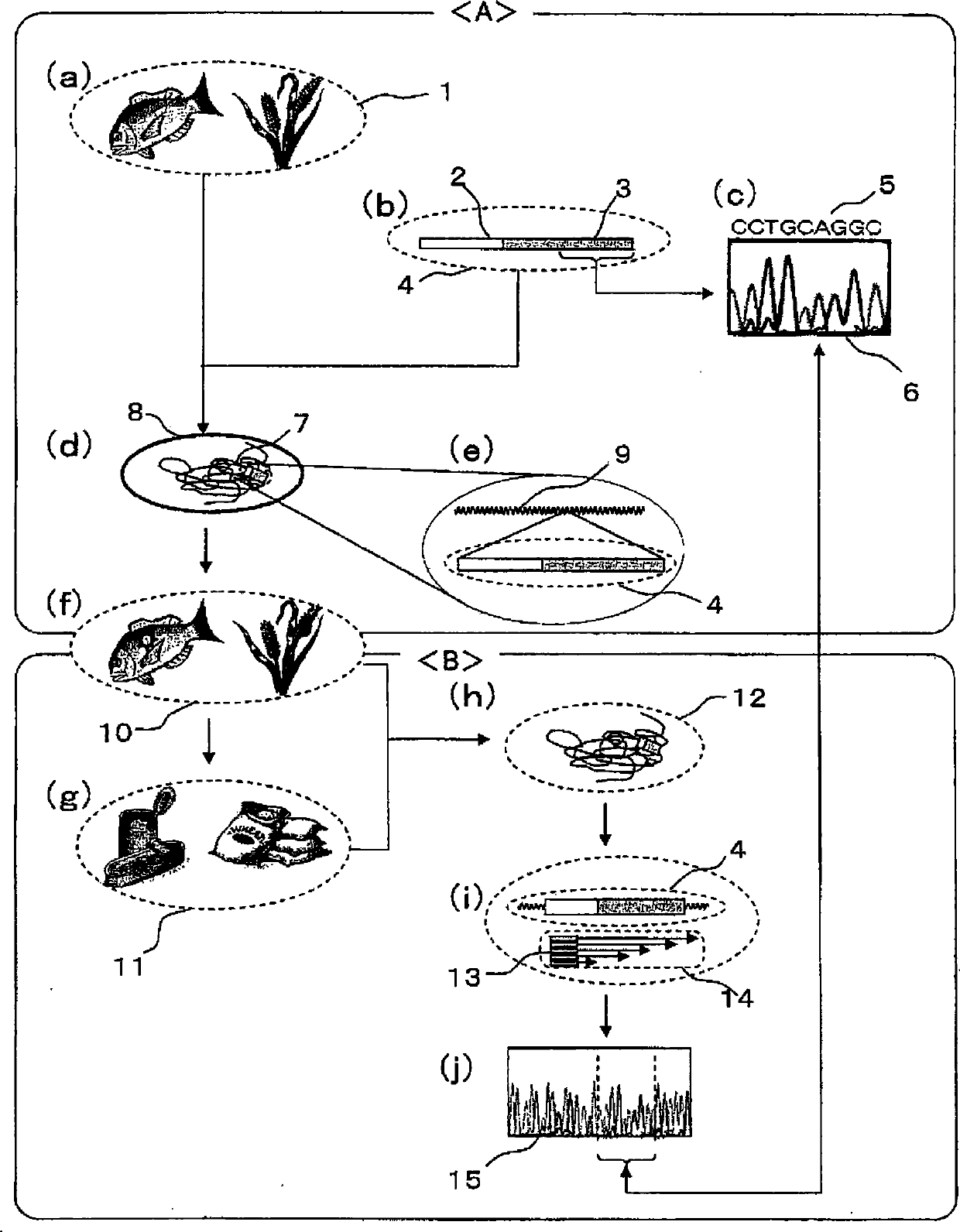
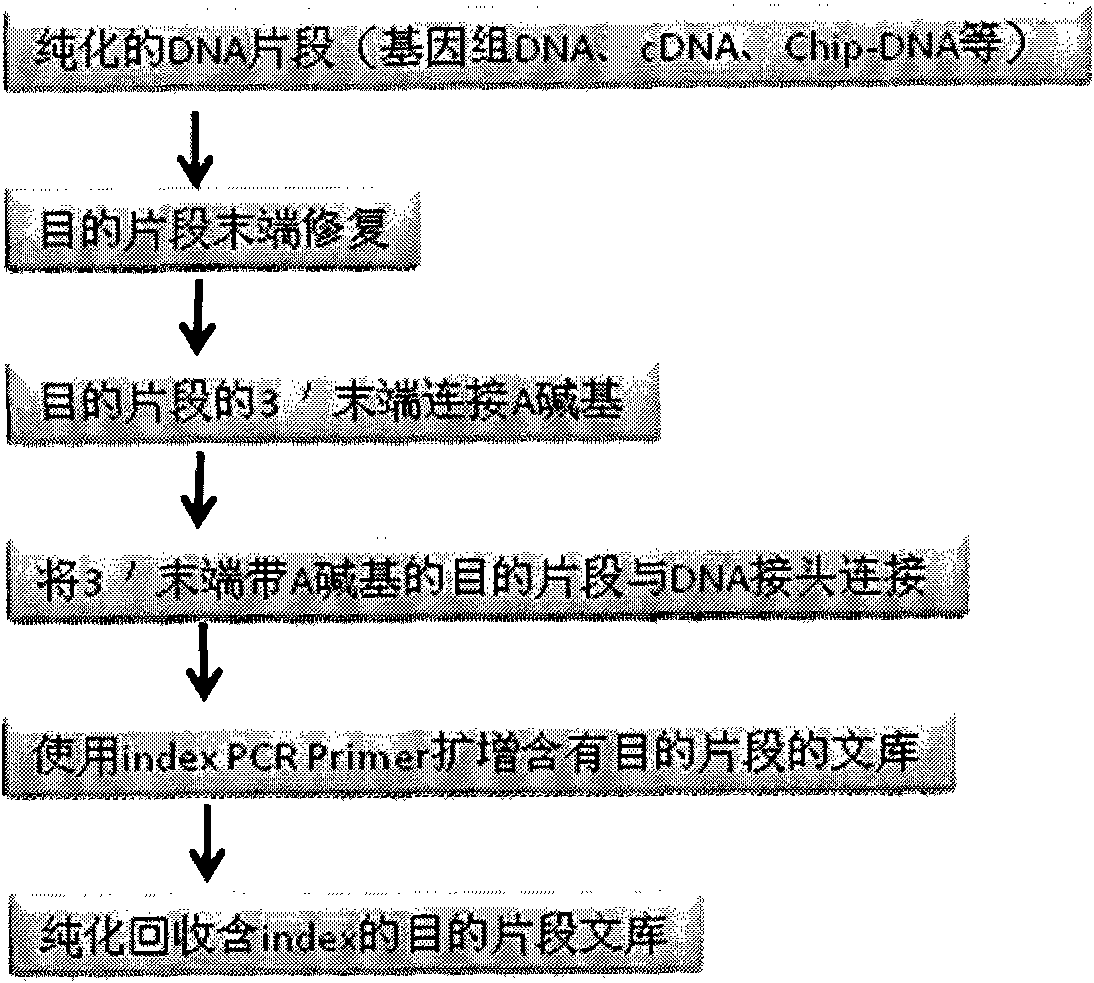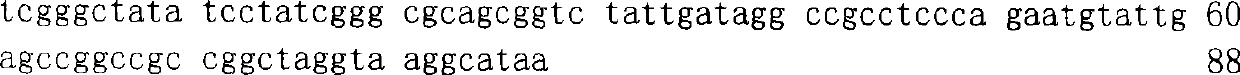Patents
Literature
76 results about "Dna labeling" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Rare cell analysis using sample splitting and DNA tags
ActiveUS20100136529A1Microbiological testing/measurementLaboratory glasswaresFetal abnormalityRare cell
The present invention provides systems, apparatuses, and methods to detect the presence of fetal cells when mixed with a population of maternal cells in a sample and to test fetal abnormalities, e.g. aneuploidy. The present invention involves labeling regions of genomic DNA in each cell in said mixed sample with different labels wherein each label is specific to each cell and quantifying the labeled regions of genomic DNA from each cell in the mixed sample. More particularly the invention involves quantifying labeled DNA polymorphisms from each cell in the mixed sample.
Owner:VERINATA HEALTH INC +2
High throughput oligonucleotide sequencing method
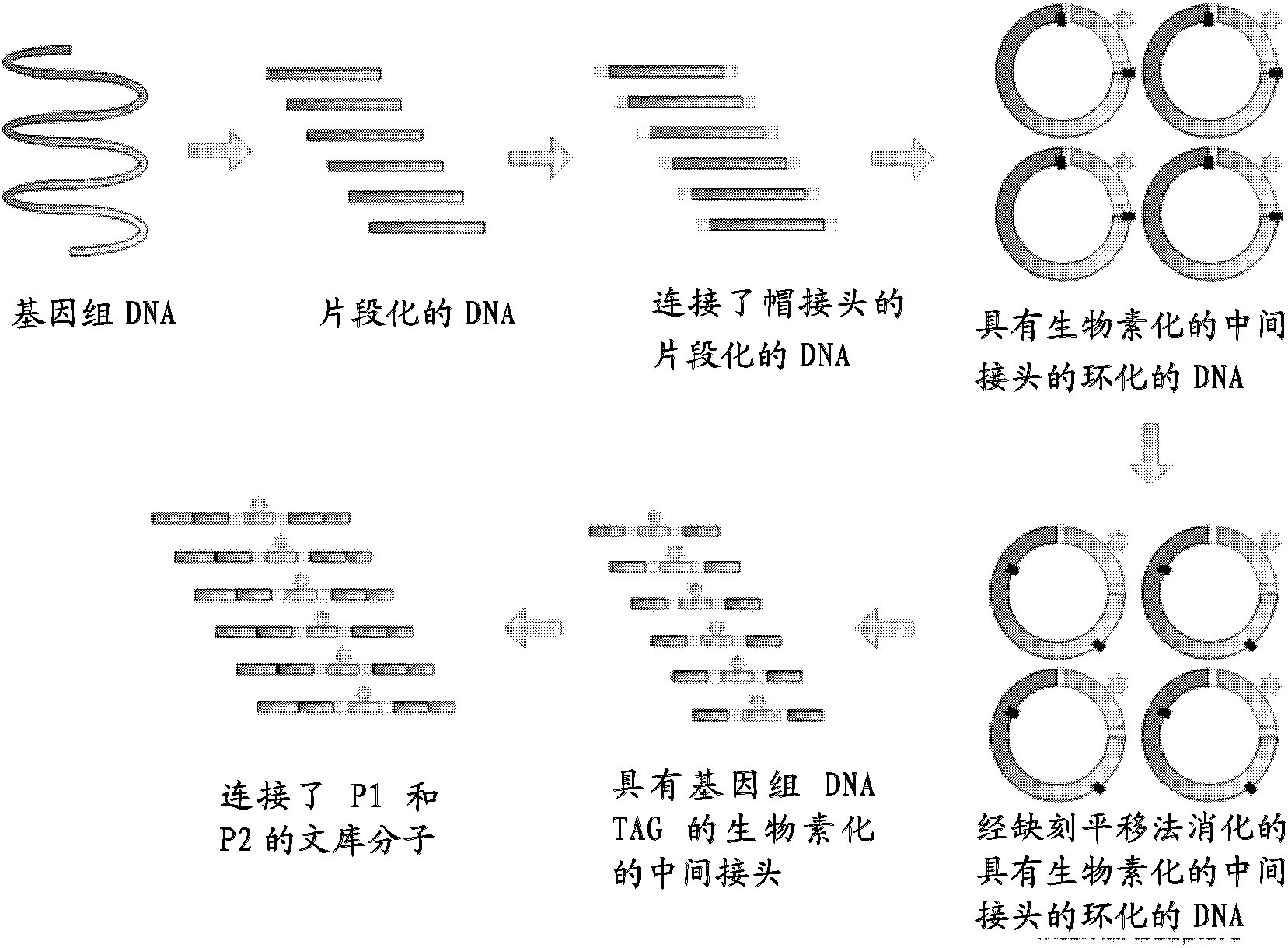
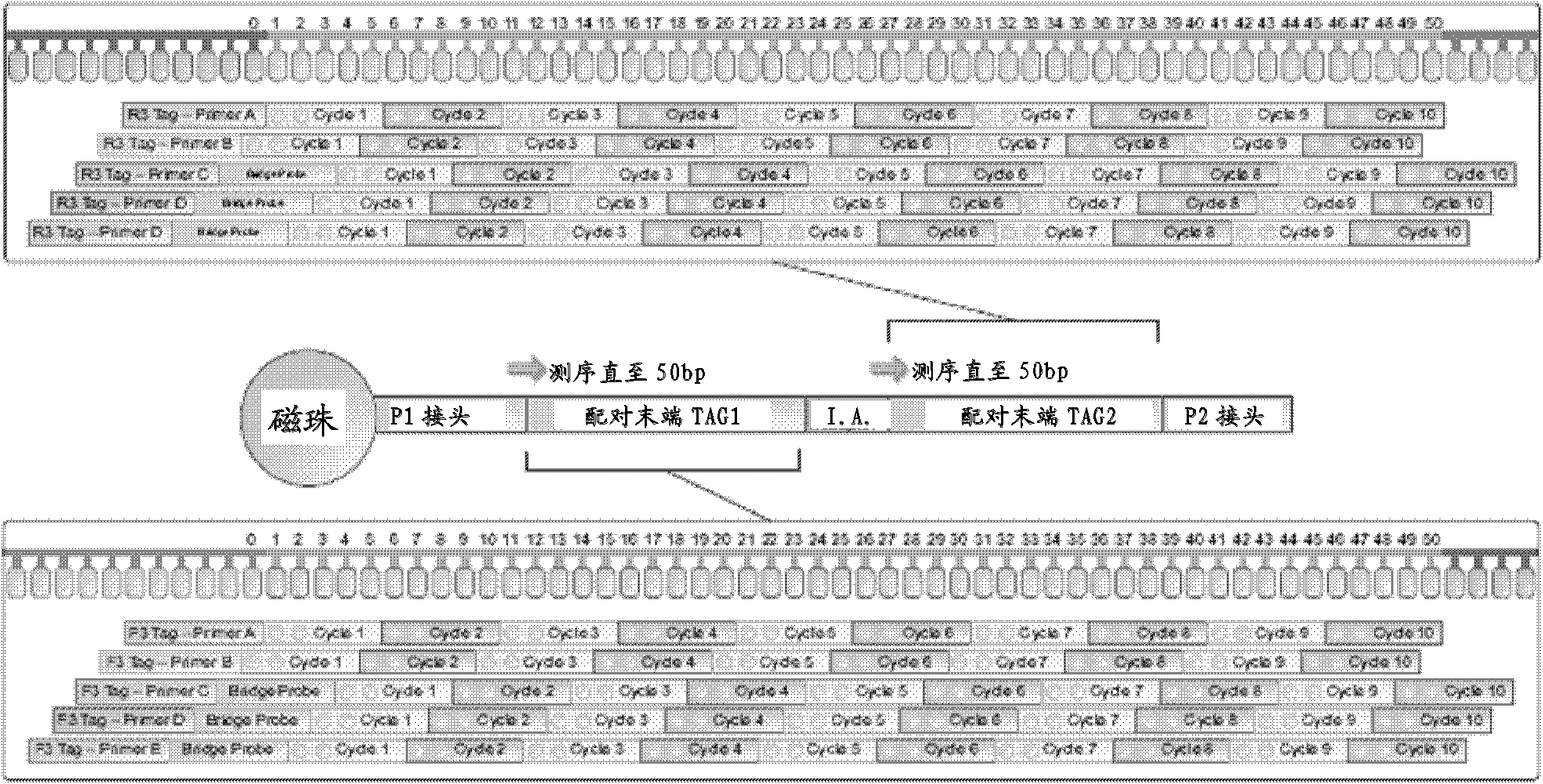
The present invention relates to the field of gene engineering, provides a high throughout oligonucleotides sequencing method. Said sequencing method includes following steps: the generation of a short DNA tag, single-molecule PCR amplification benefit-marking sequence, enrichment of high-density DNA tags and large-scale cycle parallel sequencing, etc. By using a molecular scale for parallel sequencing in a sequencing method which takes complexation as the base or expanded synthesis as the base, the scale or an expanded anchor primer is used for expanding the base points which are read out on a template. By using the primer, longer sequence on the template can be read out, and the only identification can be obtained from the genome sequence of various organisms with the range from microorganisms, plants, and animals to human, thereby having greater practical value.
Owner:SHENGZHEN CHINA GENE TECH COMPANY
Method for carrying out high-throughput sequencing on TCR (T cell receptor) or BCR (B cell receptor) and method for correcting multiplex PCR (polymerase chain reaction) primer deviation by utilizing tag sequences
InactiveCN103710454AFully functionalReduce sequencing errorsMicrobiological testing/measurementDNA/RNA fragmentationV regionPcr ctpp
The invention provides a method for carrying out high-throughput sequencing on a TCR (T cell receptor) or a BCR (B cell receptor). The method is characterized by designing upstream primers according to gene features of a V region of the TCR or the BCR and designing downstream primers according to gene features of a C region or a J region of the TCR or the BCR and obtaining sequences of the of the TCR or the BCR in combination with the multiplex PCR (polymerase chain reaction) technology and high-throughput sequencing, thus analyzing the rearrangement information of the TCR or the BCR. Compared with 25-30 cycles of existing multiplex PCR, two cycles of the multiplex PCR technology provided by the invention can conduce to greatly reducing the sequencing errors caused by primer amplification preference. Besides, the invention also provides a method for correcting multiplex PCR (polymerase chain reaction) primer deviation by utilizing DNA (deoxyribonucleic acid) tag sequences, thus further reducing the sequencing errors caused by primer amplification preference and intrinsic sequencing errors of high-throughput sequencing.
Owner:SOUTH UNIVERSITY OF SCIENCE AND TECHNOLOGY OF CHINA +1
DNA index library building method based on high throughput sequencing
ActiveCN102409048AImprove production efficiencyHigh sequencing throughputNucleotide librariesMicrobiological testing/measurementSingle sampleA-DNA
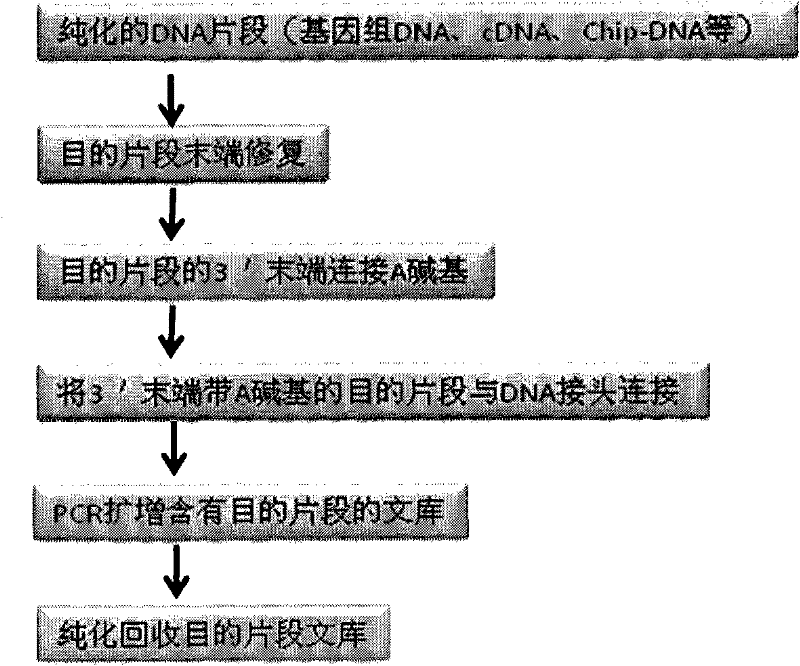
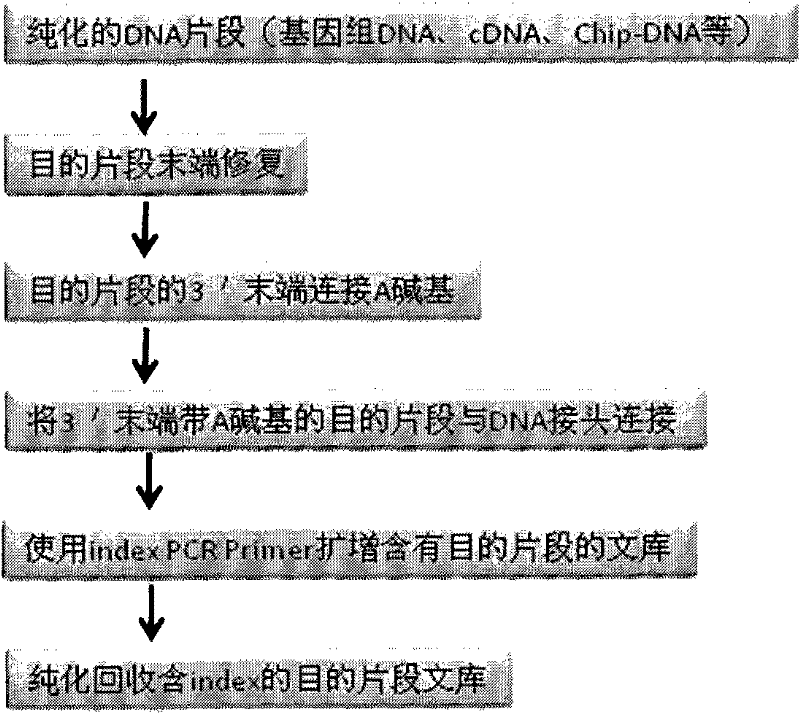
The invention provides unique index sequences with a length of 7bp. The index sequences can be respectively imported into a DNA(deoxyribonucleic acid) index library through adapter link and PCR (polymerase chain reaction). The invention provides a method for building the DNA index library by using the index sequences based on a solexa sequencing platform of the current illumina company, and the method is applied to solexa DNA sequencing and has the effects on improving the preparation efficiency of the DNA index library, increasing the sequencing throughput of the DNA samples and lowering thesolexa sequencing cost of a single sample.
Owner:BGI TECH SOLUTIONS
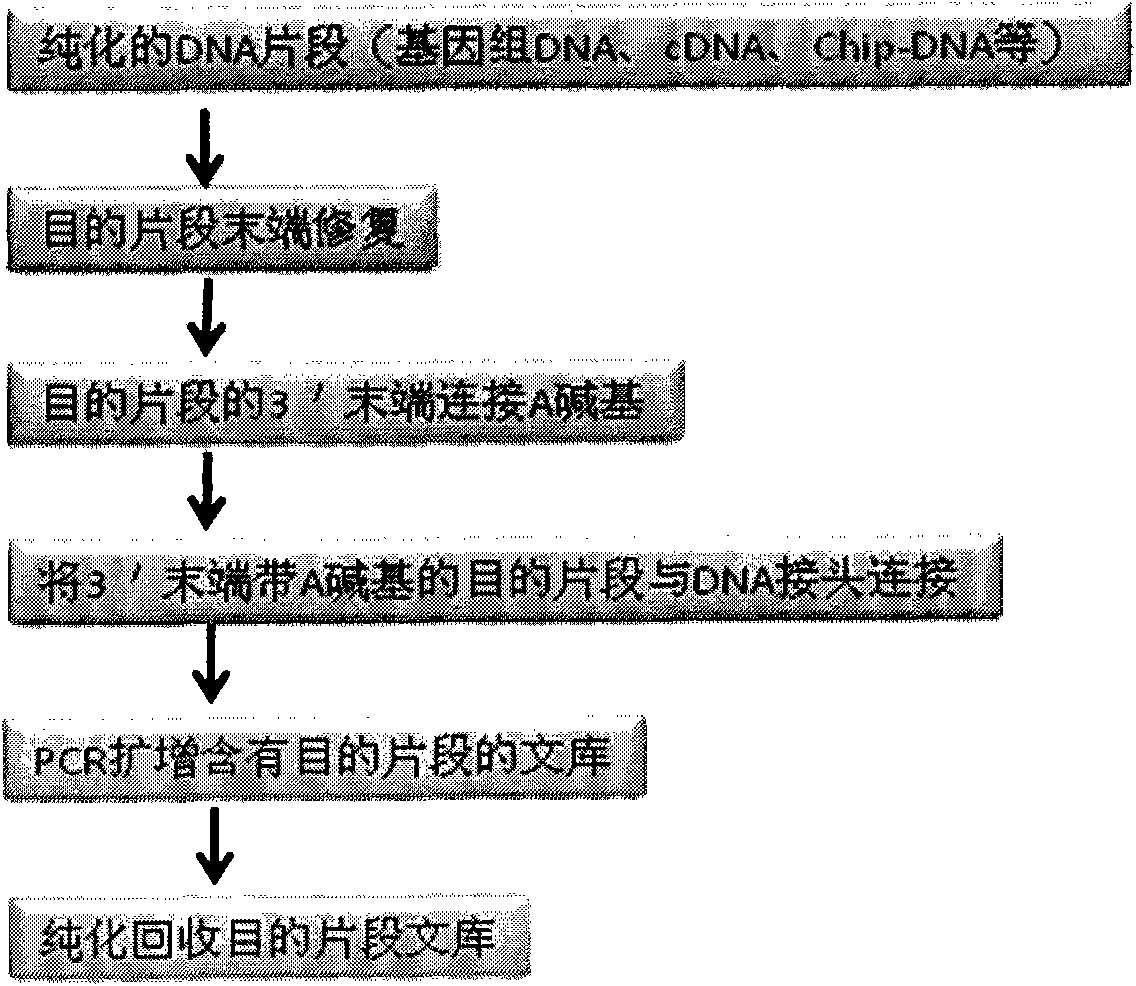
Joint connection-based deoxyribonucleic acid (DNA) polymerase chain reaction (PCR)-free tag library construction method
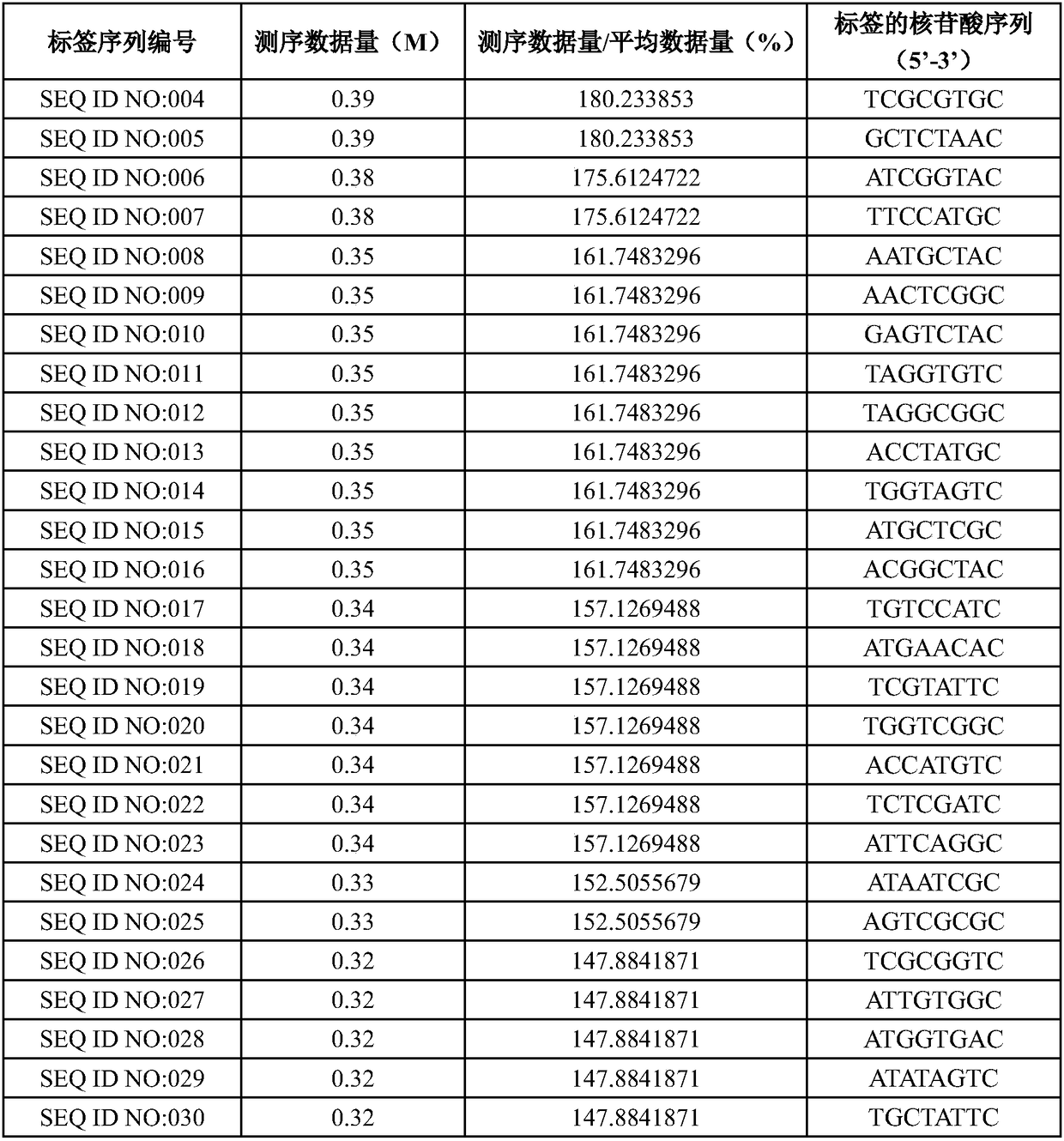
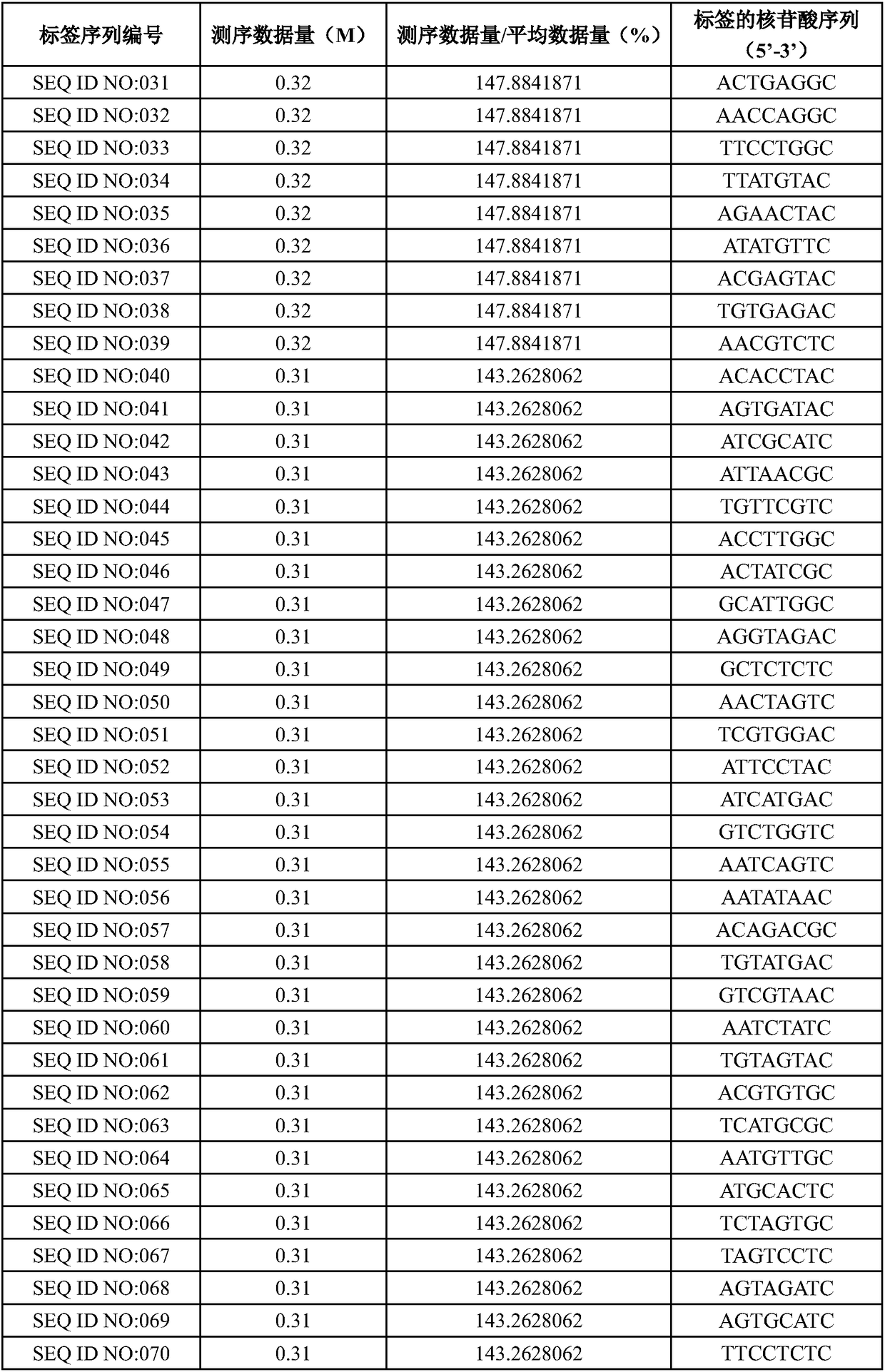
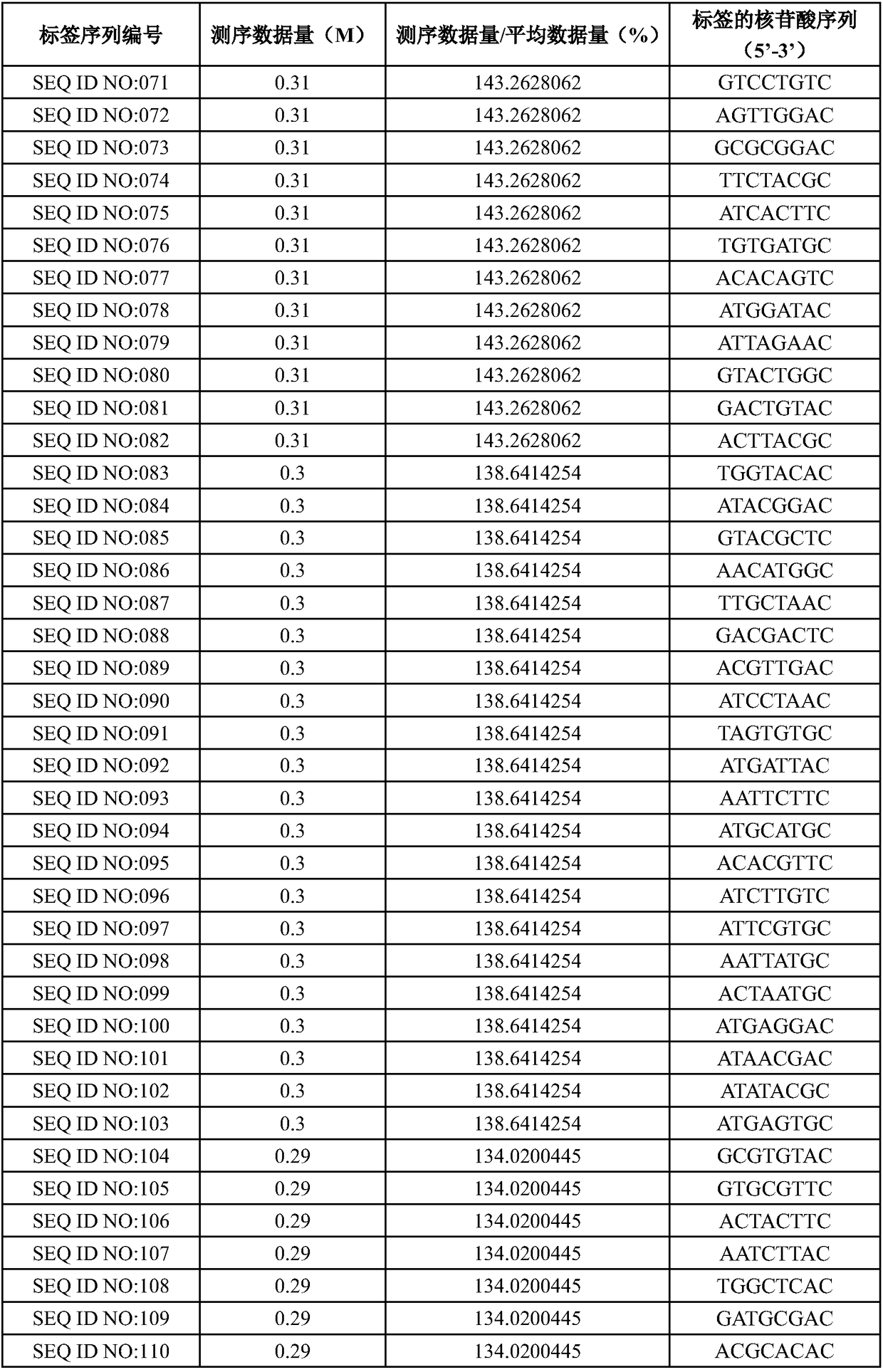
The invention designs 161 unique tag sequences with the lengths of 8 bp. Tags are embedded into a deoxyribonucleic acid (DNA) joint so as to form a DNA polymerase chain reaction (PCR)-free tag joint. The invention also provides a DNA tag library which introduces the tag sequence by connecting the DNA PCR-free tag joint without a PCR and is applied to solexa DNA sequencing. After a library construction method is optimized, the DNA tag library is constructed without the PCR.
Owner:BGI TECH SOLUTIONS
DNA labels, PCR primers and application thereof
ActiveCN104131008AMicrobiological testing/measurementLibrary creationNucleotideNucleic acid sequencing
The invention discloses DNA labels, PCR primers and application thereof. A group of the DNA labels are composed of nucleotides as shown in SEQ ID No. 79 to 129. The group of the DNA labels can be used for establishment of a nucleic acid sequencing library to realize accurate differentiation of the nucleic acid sequencing library. The sources of DNA samples can be accurately characterized by connecting the DNA labels with DNA or equivalents thereof.
Owner:海南华大基因科技有限公司
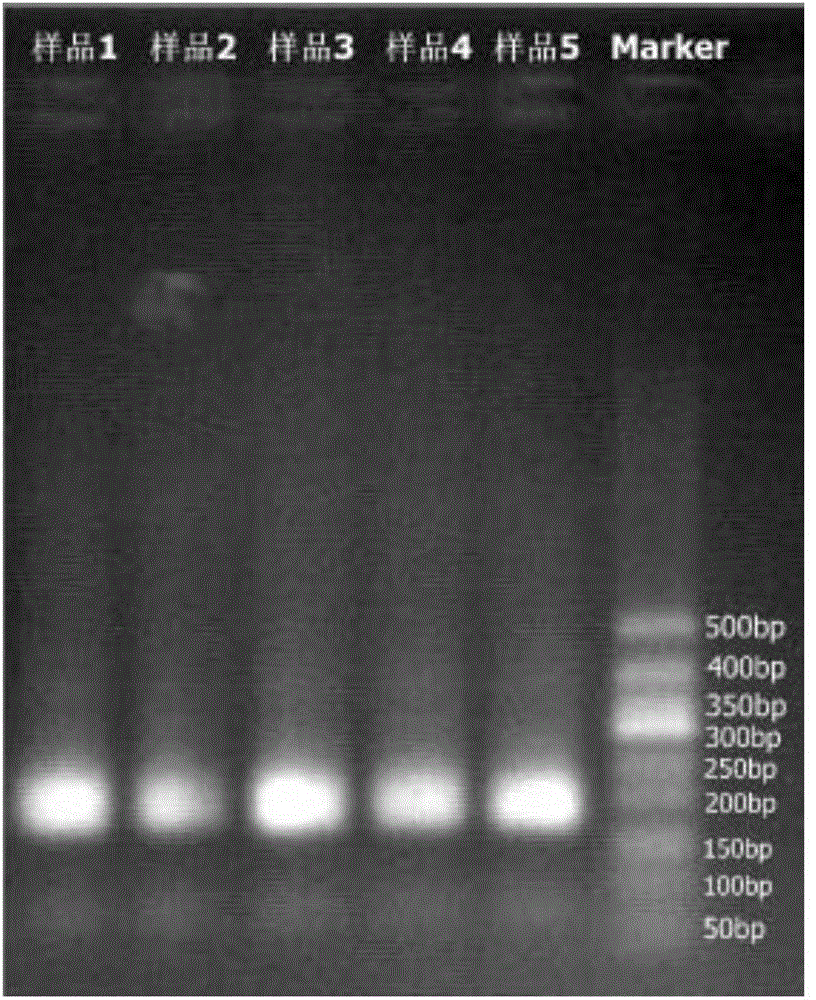
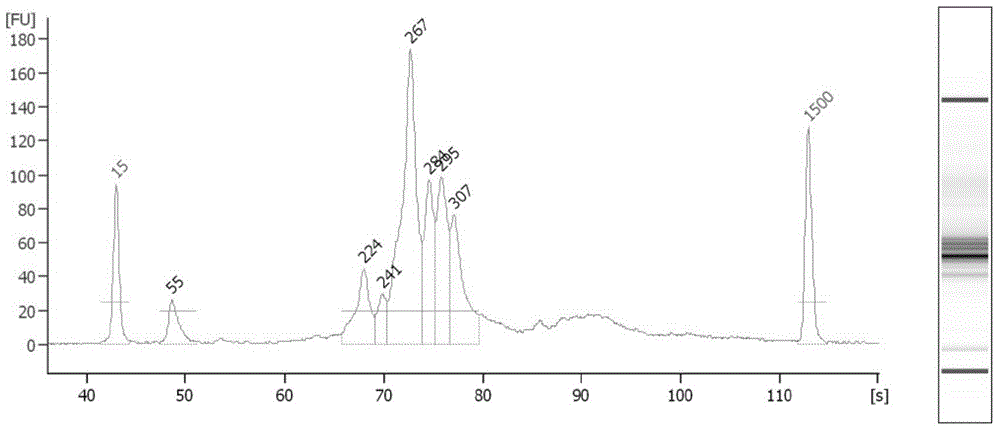
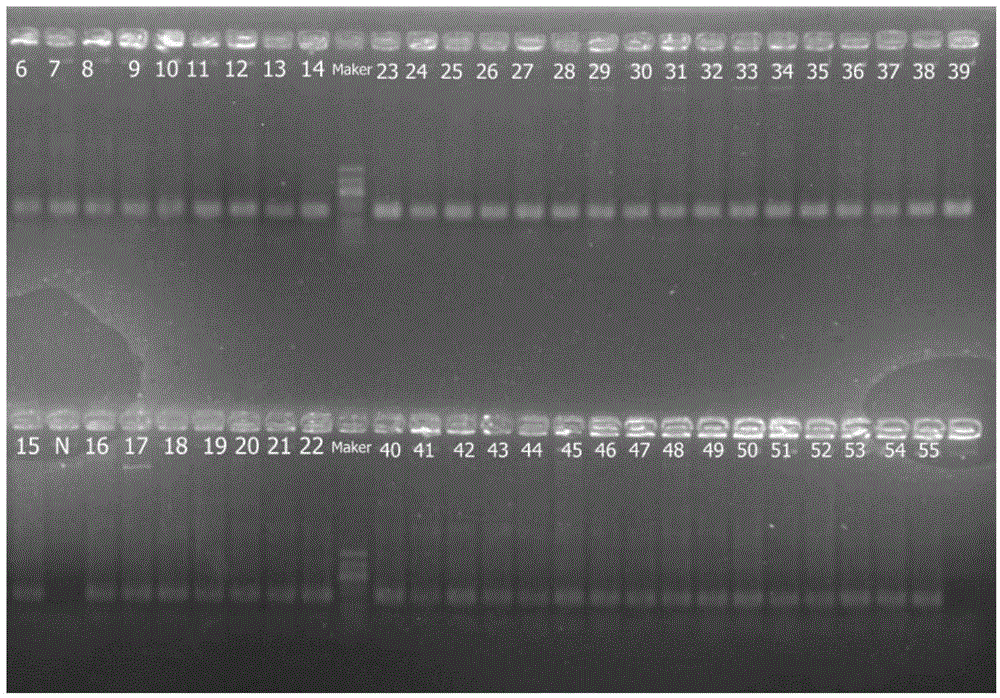
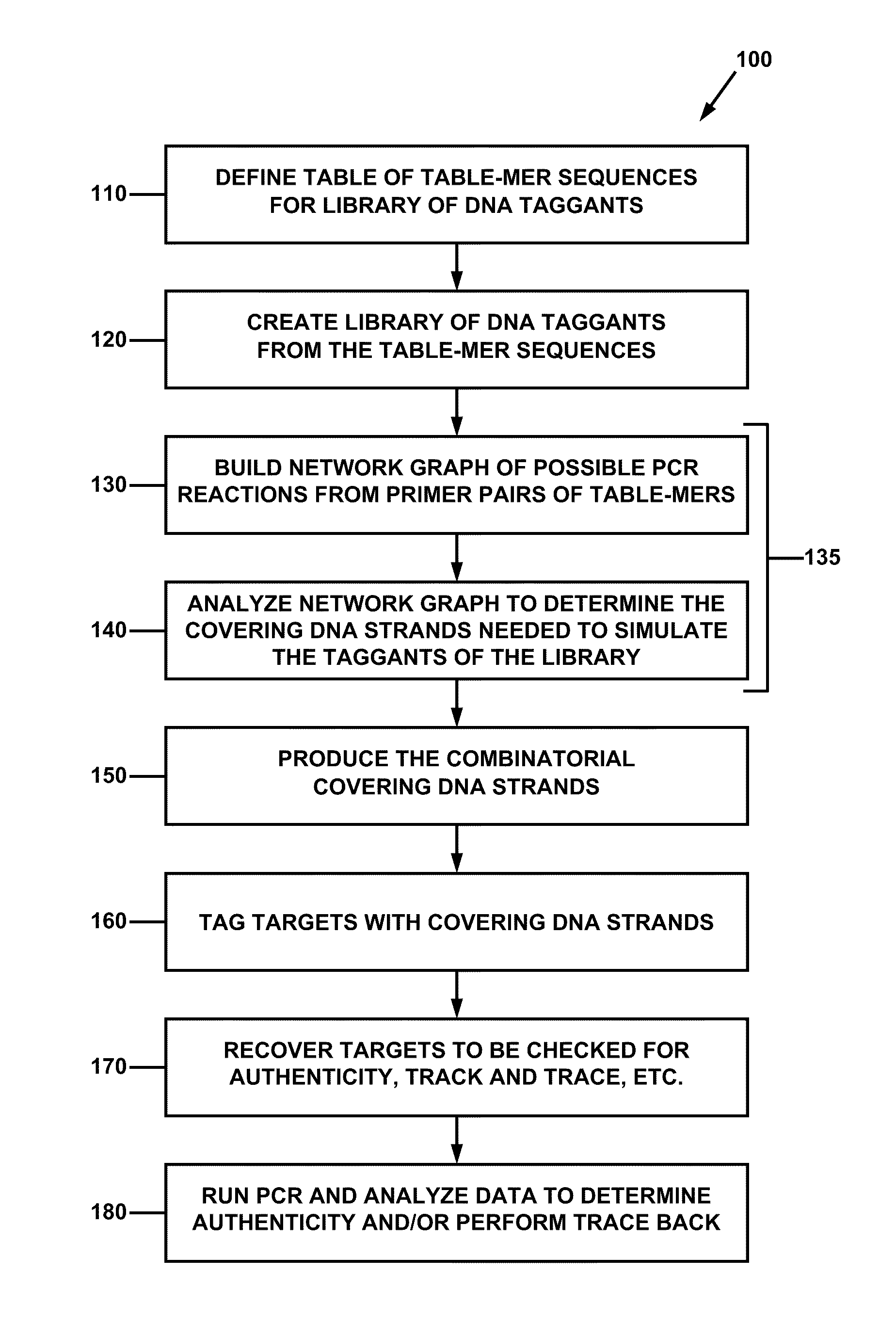
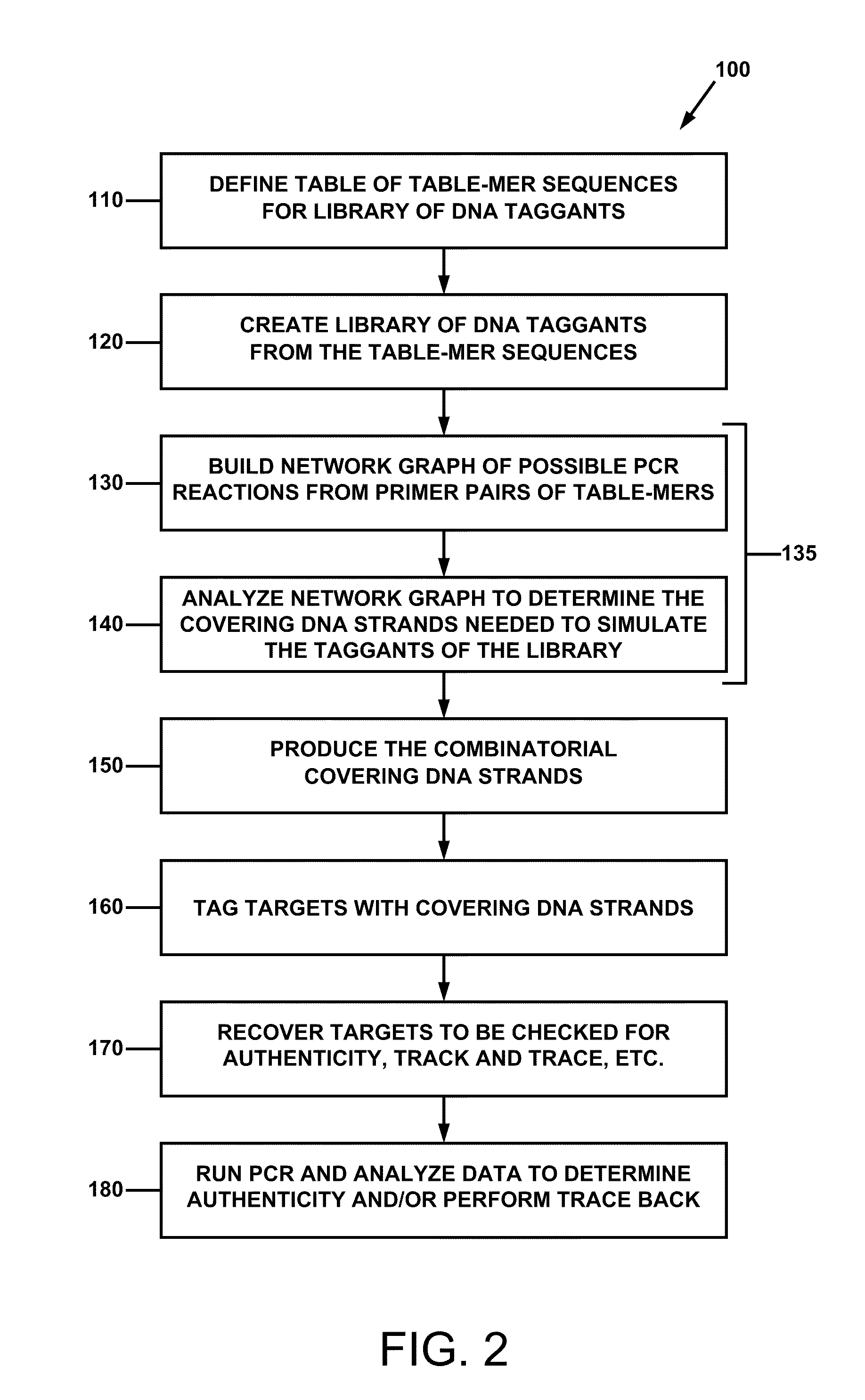
Combinatorial DNA taggants and methods of preparation and use thereof
ActiveUS20110165569A1Prevent counterfeitingAuthentication processSugar derivativesMicrobiological testing/measurementNucleotide sequencingBiology
DNA taggants in which the nucleotide sequences are defined according to combinatorial mathematical principles. Methods of defining nucleotide sequences of the combinatorial DNA taggants, and using such taggants for authentication and tracking and tracing an object or process are also disclosed.
Owner:PATENT INNOVATIONS
Porphyra yezoensis microsatellite marker screening method and use thereof
InactiveCN101550434AImprove reliabilityHigh yieldMicrobiological testing/measurementFermentationResource protectionGermplasm
The invention belongs to molecular biology DNA labeling technique and application field, concretely relates to a porphyra yezoensis microsatellite marker screening method, and a method for analyzing germplasm genetic diversity by the makers, thereby lay foundations for genetic structure analysis of porphyra yezoensis, germplasm resource protection and molecular marker auxiliary breeding.
Owner:OCEAN UNIV OF CHINA
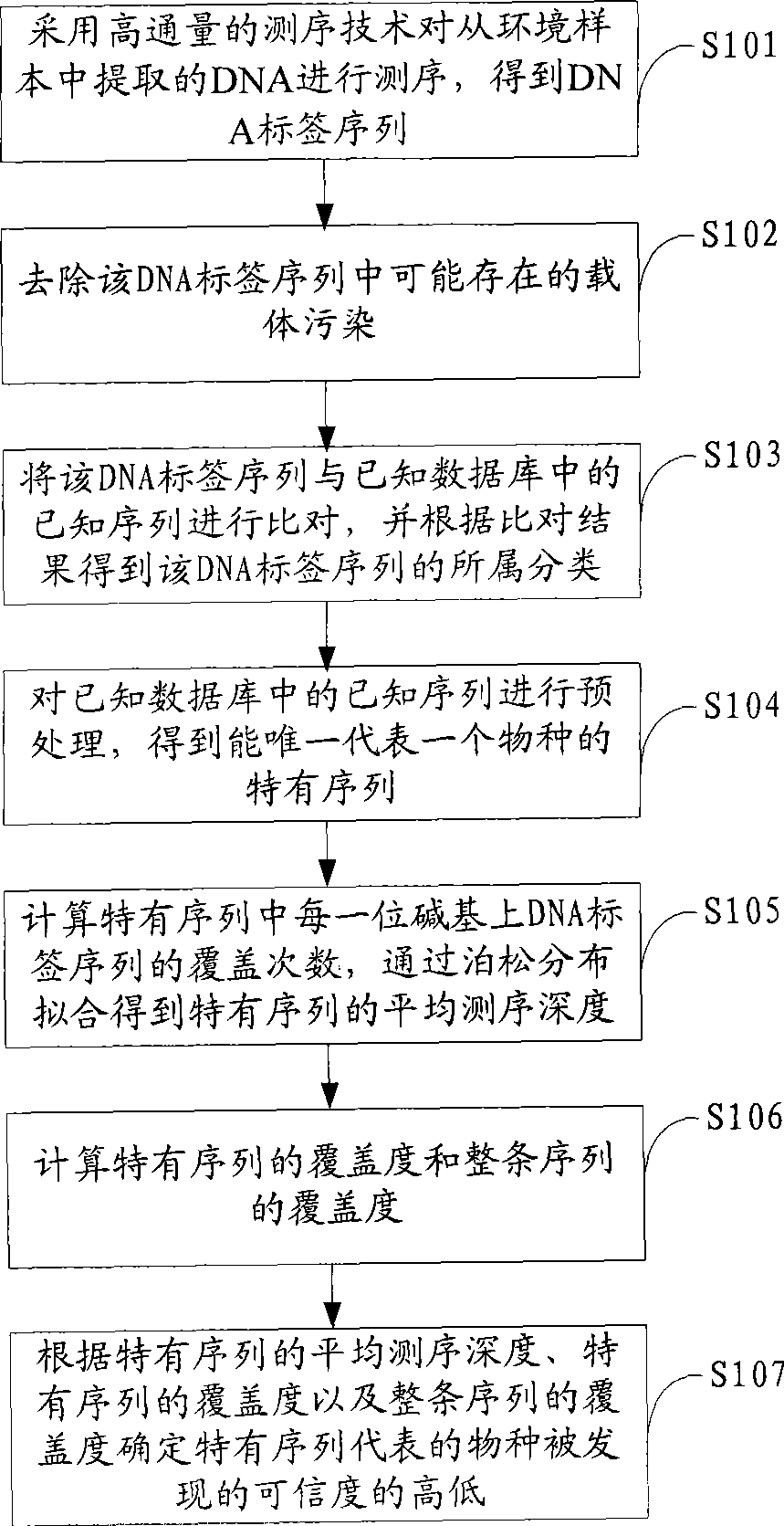
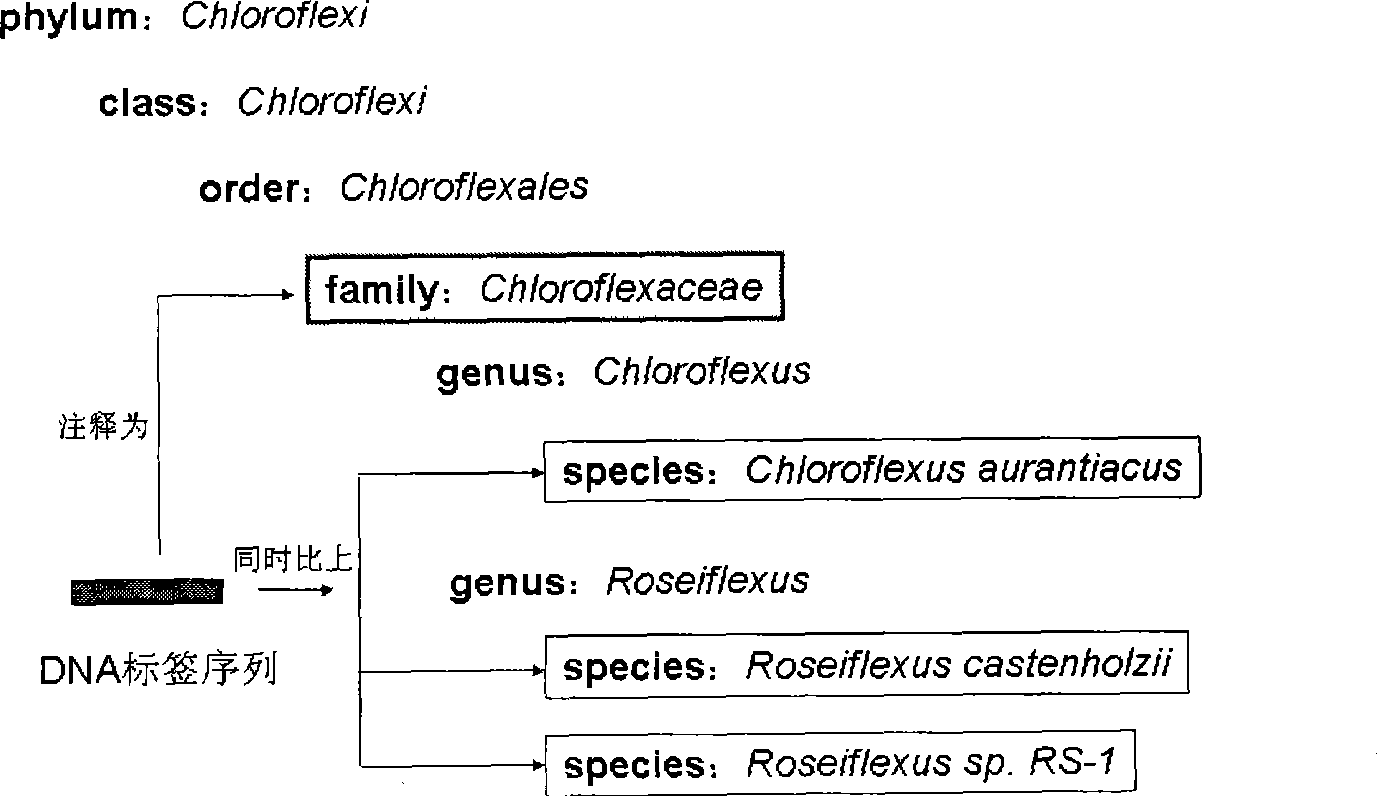
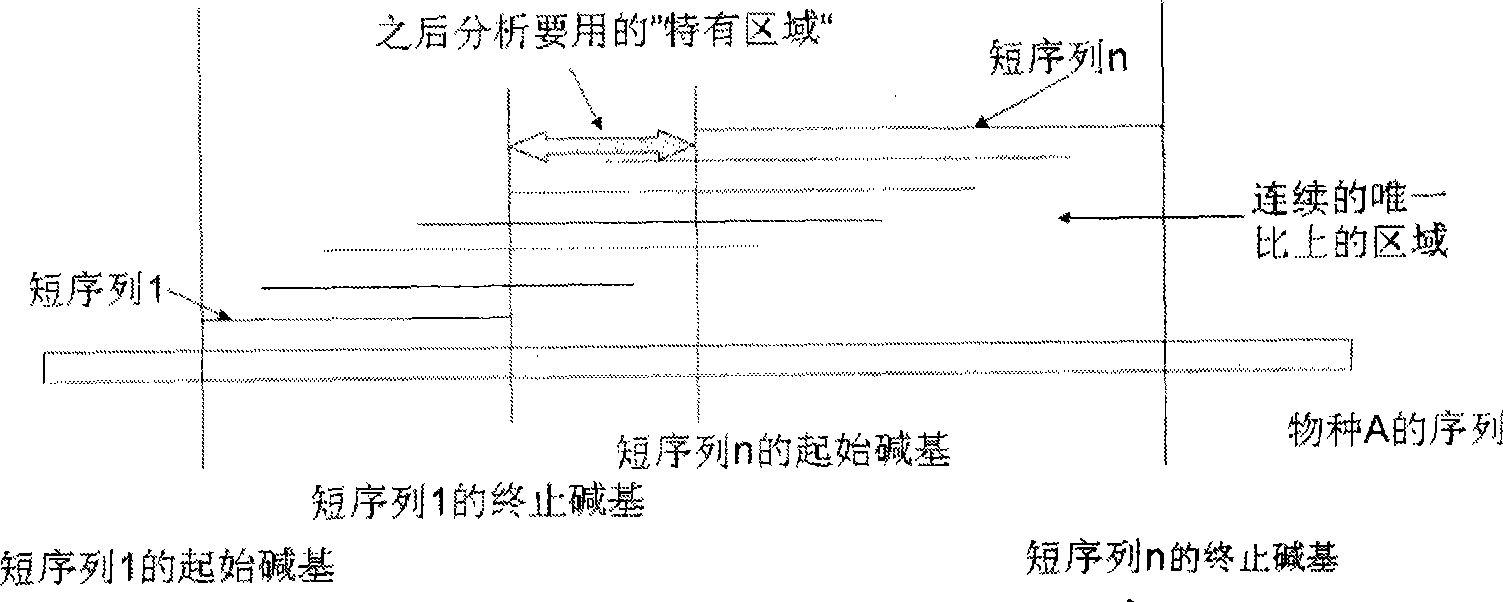
Environmental microorganism detection method and system
ActiveCN101748213AComprehensive sequencingEfficient identificationBioreactor/fermenter combinationsBiological substance pretreatmentsMicroorganismA-DNA
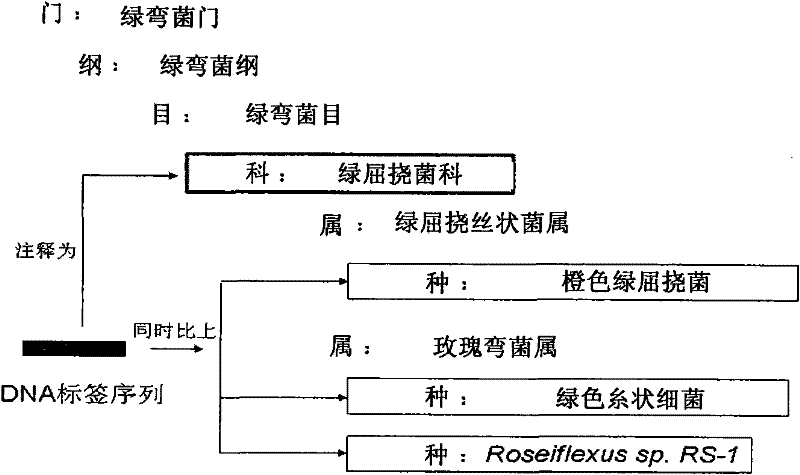
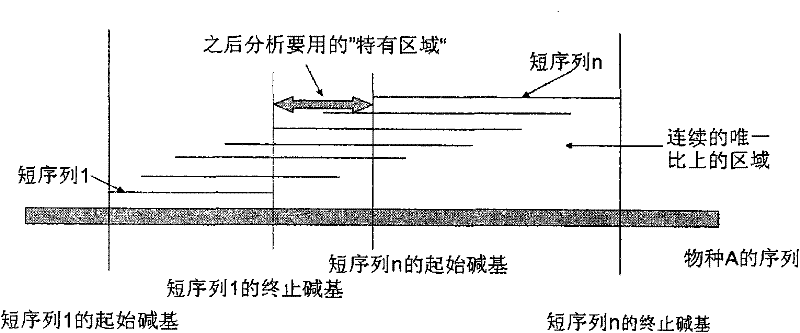
The invention is applicable to the field of biological engineering, and provides an environmental microorganism detection method and a system thereof. The method comprises the following steps: sequencing the DNA extracted from environment samples by the high throughput sequencing technology to obtain a DNA tag sequence; eliminating carrier pollution existing in DNA tag sequence; comparing the DNAtag sequence with the known sequence in the known database, and determining the category to which the DNA tag sequence belongs according to the comparison result. The embodiment of the invention can detect which microorganism species or which class of microorganism species possibly exists in environment samples.
Owner:SHENZHEN HUADA GENE INST +1
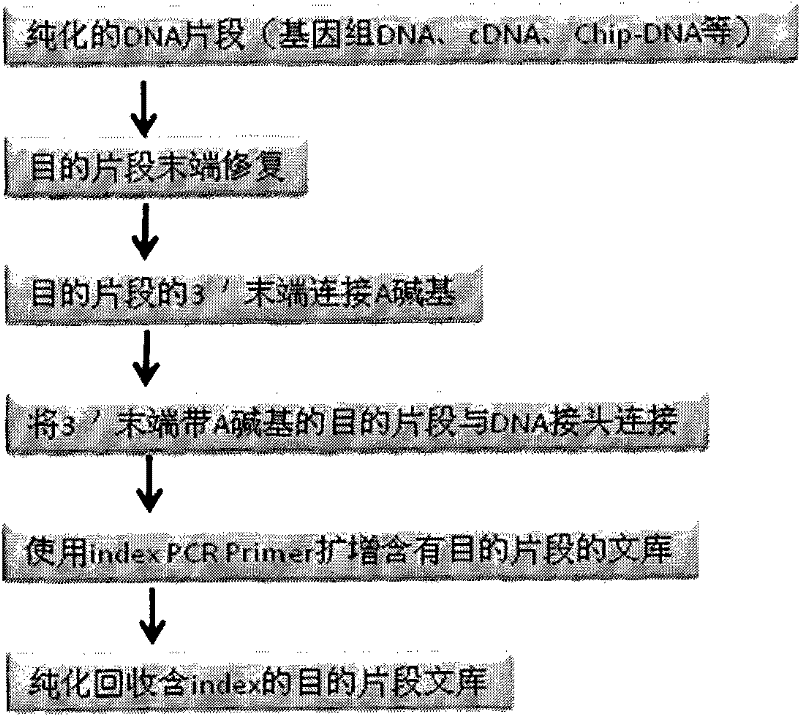
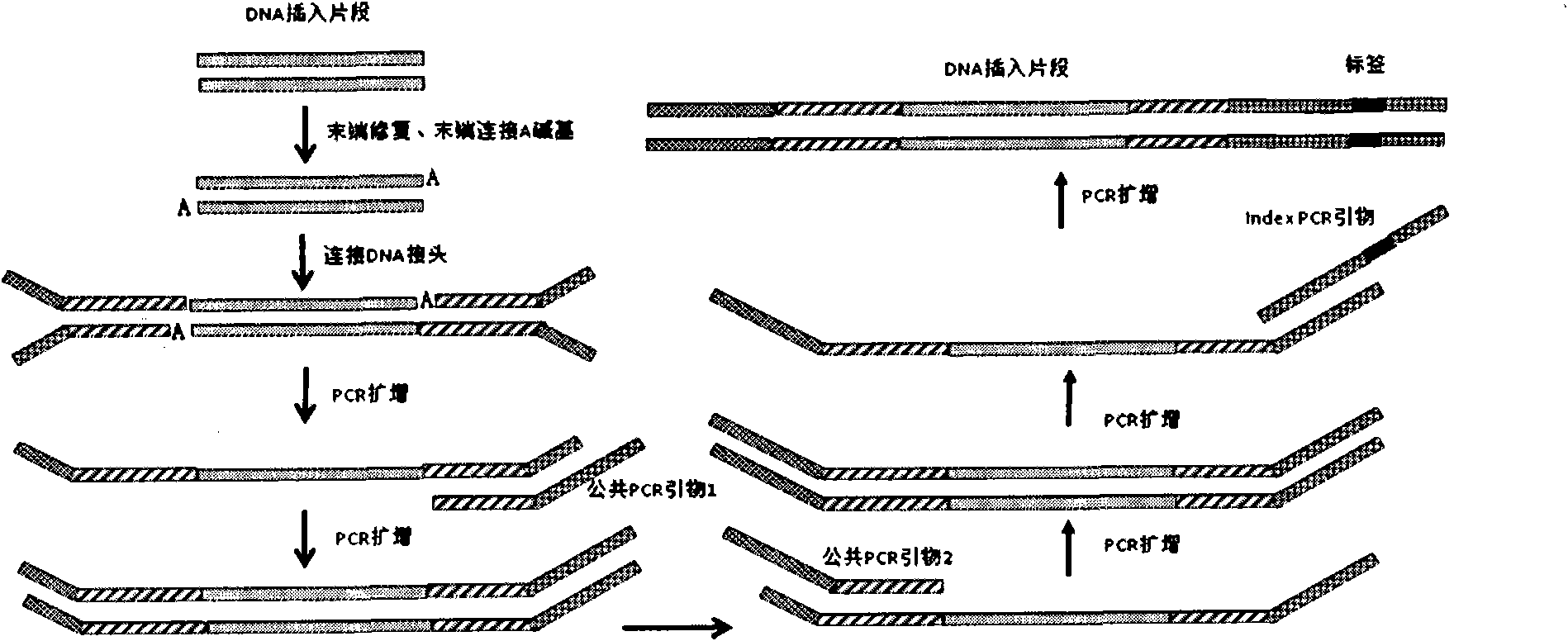
DNA(deoxyribonucleic acid) index library building method based on PCR (polymerase chain reaction)
ActiveCN102409049AImprove production efficiencyHigh sequencing throughputNucleotide librariesMicrobiological testing/measurementBiotechnologyInverse polymerase chain reaction
The invention designs 161 unique index sequences with a length of 8bp, and the indexes are embedded into DNA PCR (deoxyribonucleic acid polymerase chain reaction) primers to form DNA PCR index primers, thus being capable of importing the index sequences through PCR. The invention successfully establishes a method for building a DNA index library, and the method is applied to solexa DNA sequencing.
Owner:BGI TECH SOLUTIONS
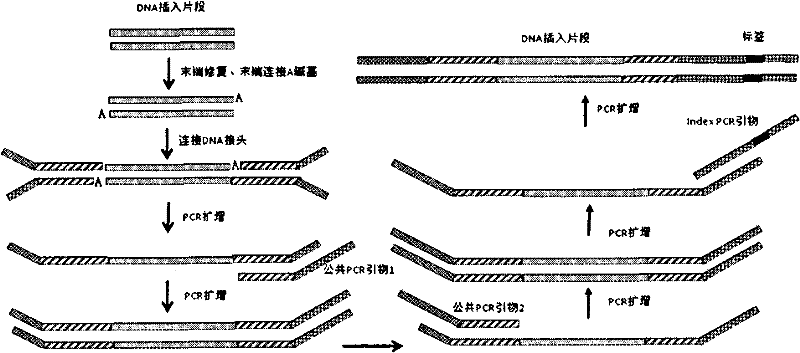
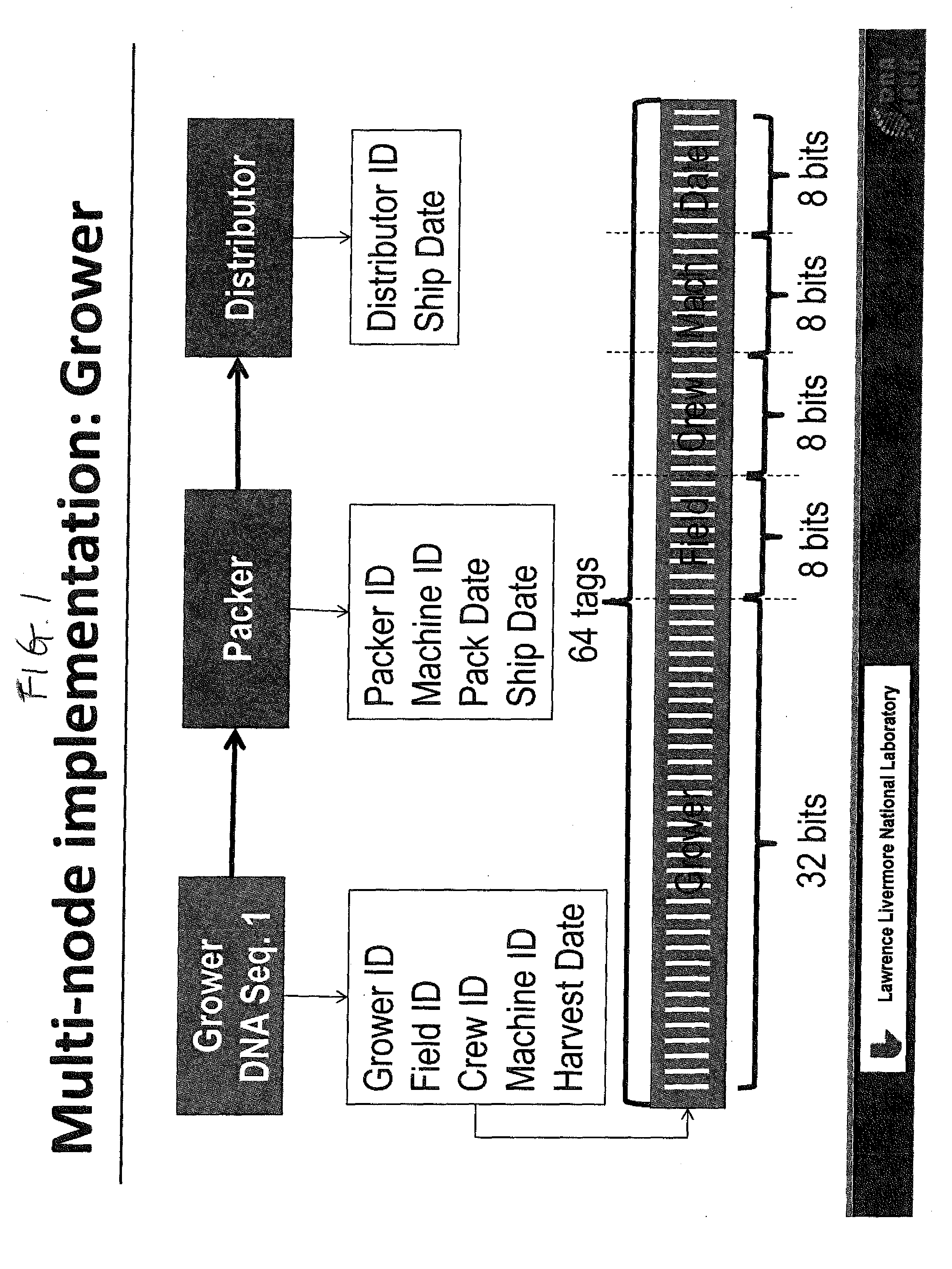
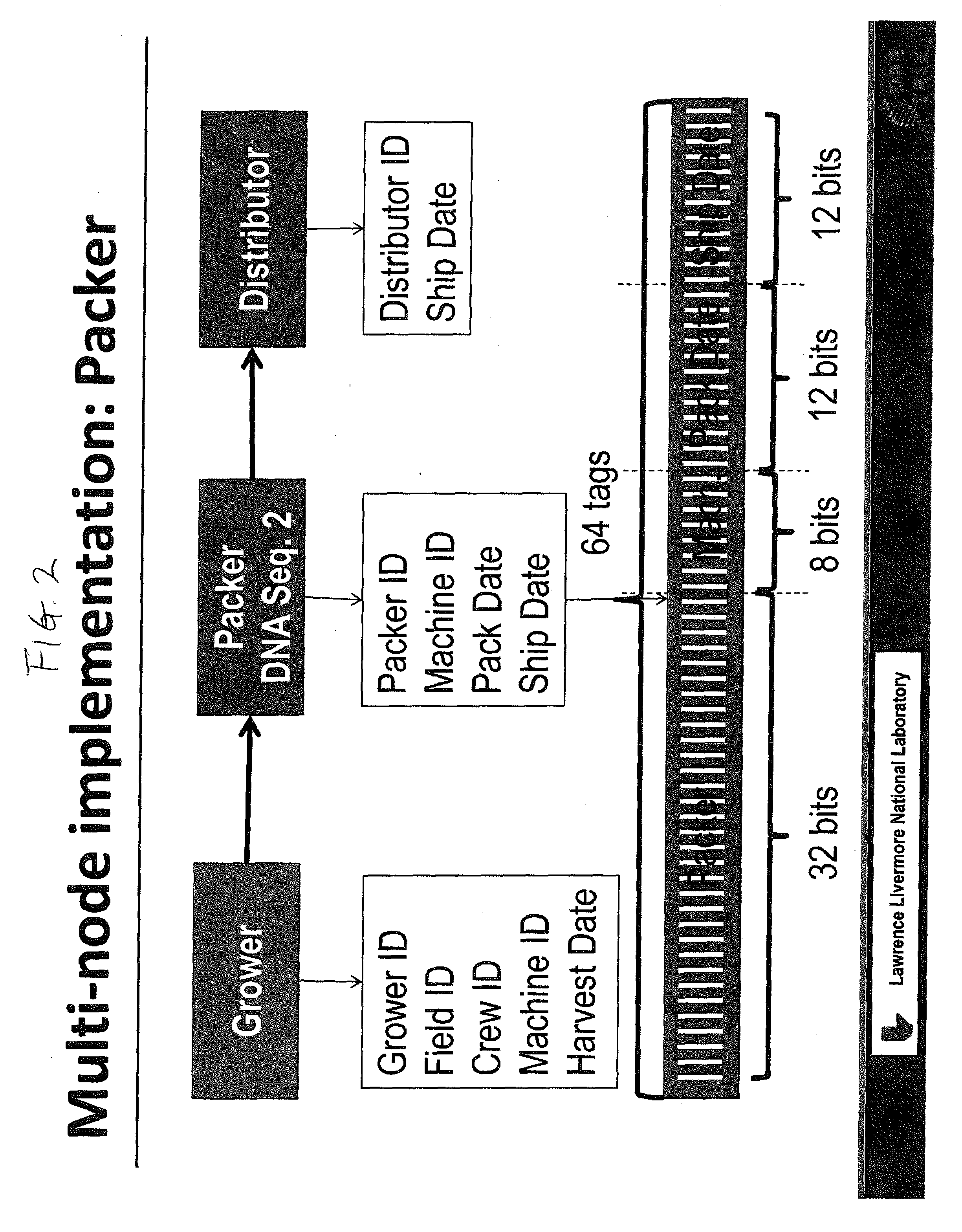
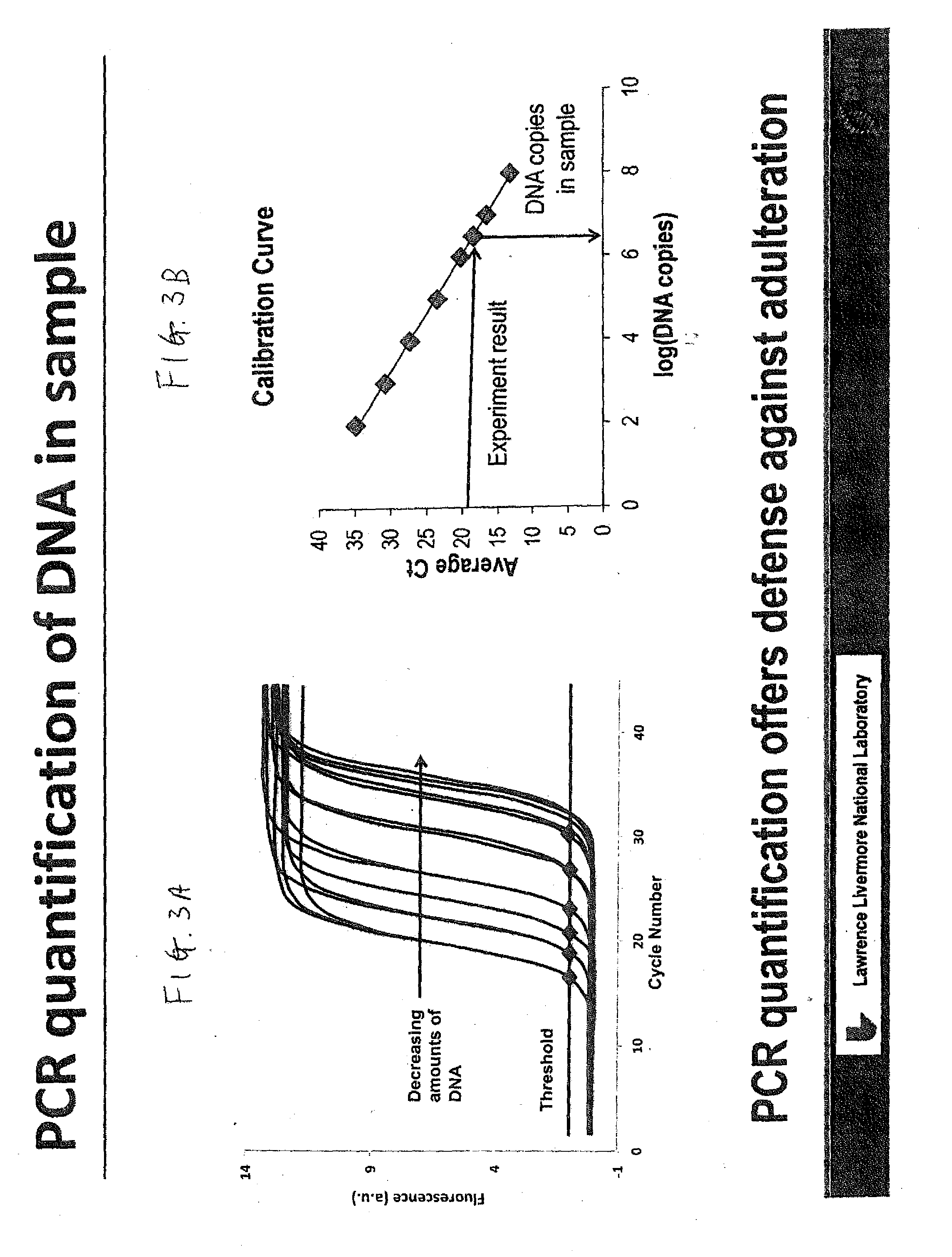
DNA Based Bar Code for Improved Food Traceability
Food distributed to consumers through a distribution chain may be traced by tagging the food with DNA tags that identify the origin of the food, such as the grower, packer and other points of distribution, and their attributes. This makes it much quicker and easier to trace the food in case of food contamination or adulteration. Preferably these attributes indicate the field, location, crew and machine used to grow and process the food, and the dates of the various steps of food harvesting, processing and distribution. Natural or synthetic DNA pieces may be used to tag items, including food items. Multidigit binary or other types of bar codes may be represented by multiple types of DNA. Each digit of the bar code may be represented by one, two or more unique DNA pieces.
Owner:LAWRENCE LIVERMORE NAT SECURITY LLC +1
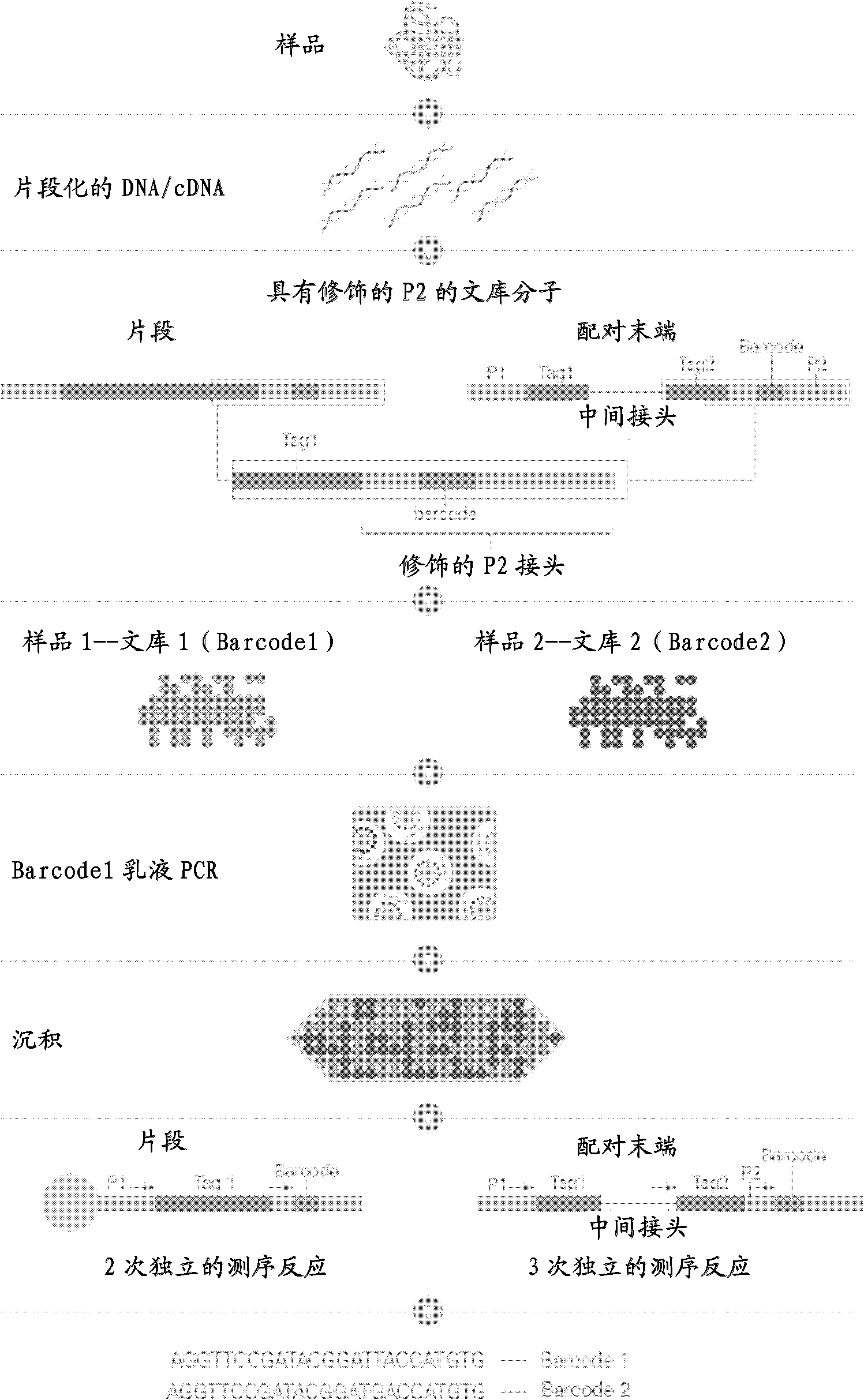
DNA index and application thereof to construction and sequencing of mate-paired indexed library
ActiveCN102690809AEnabling Hybrid SequencingAccelerated high-throughput sequencingNucleotide librariesMicrobiological testing/measurementComputational biologyMicroarray
The invention provides a set of DNA index and application thereof to construction and sequencing of mate-paired indexed library, and the DNA index has a sequence selected from SEQ ID NO:1-24. The invention also provides a method for construction and sequencing of the mate-paired index library. The method employs two independent sequencing reactions to realize mixed sequencing on a plurality of mate-paired indexed libraries in a singe sequencing chip, so as to accelerate high flux sequencing, and reduce time, reagent cost and output cost of unit data.
Owner:WUXI QINGLAN BIOLOGICAL SCI & TECH +1
Expression of proteins in E.coli
Owner:NOVO NORDISK AS
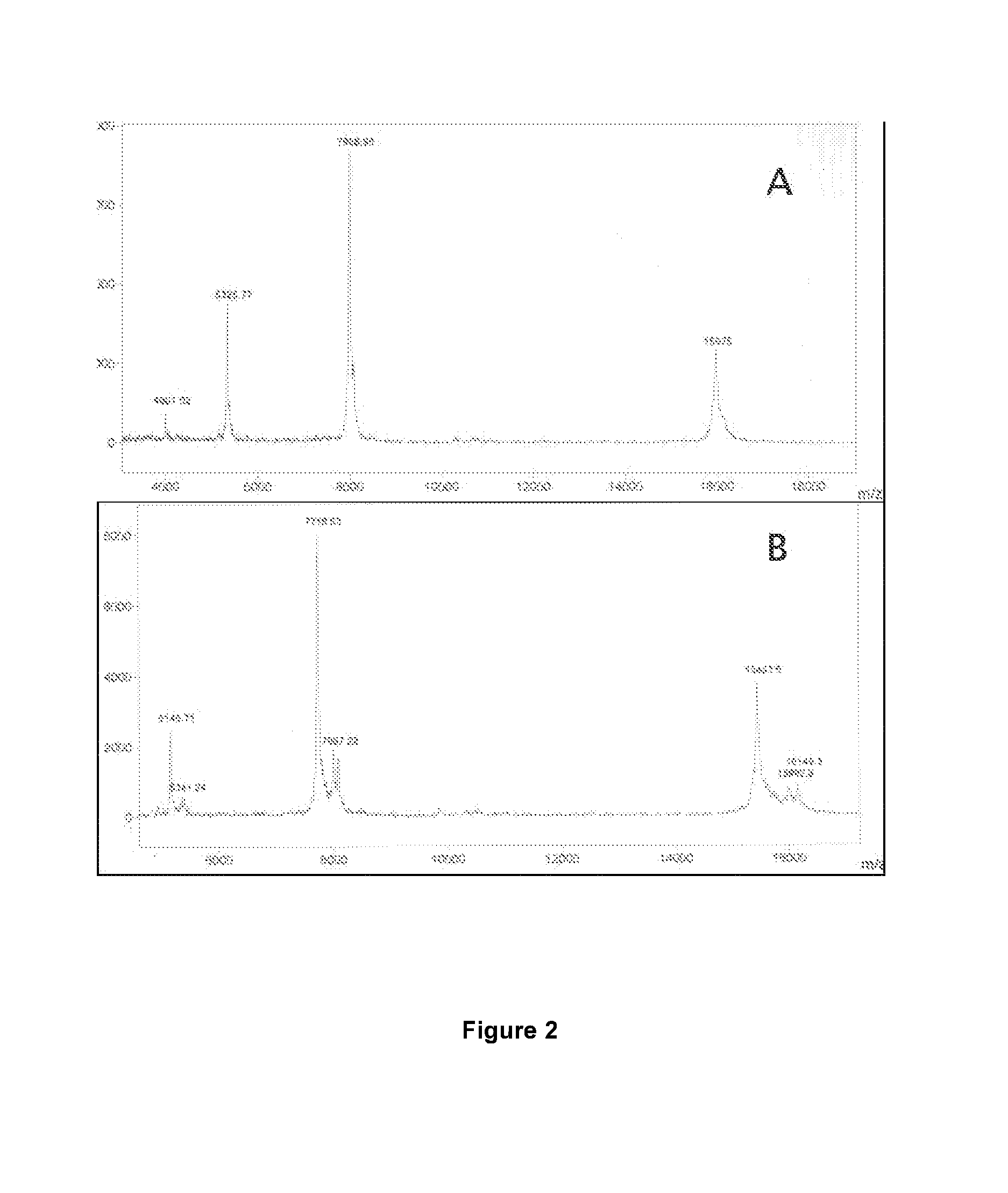
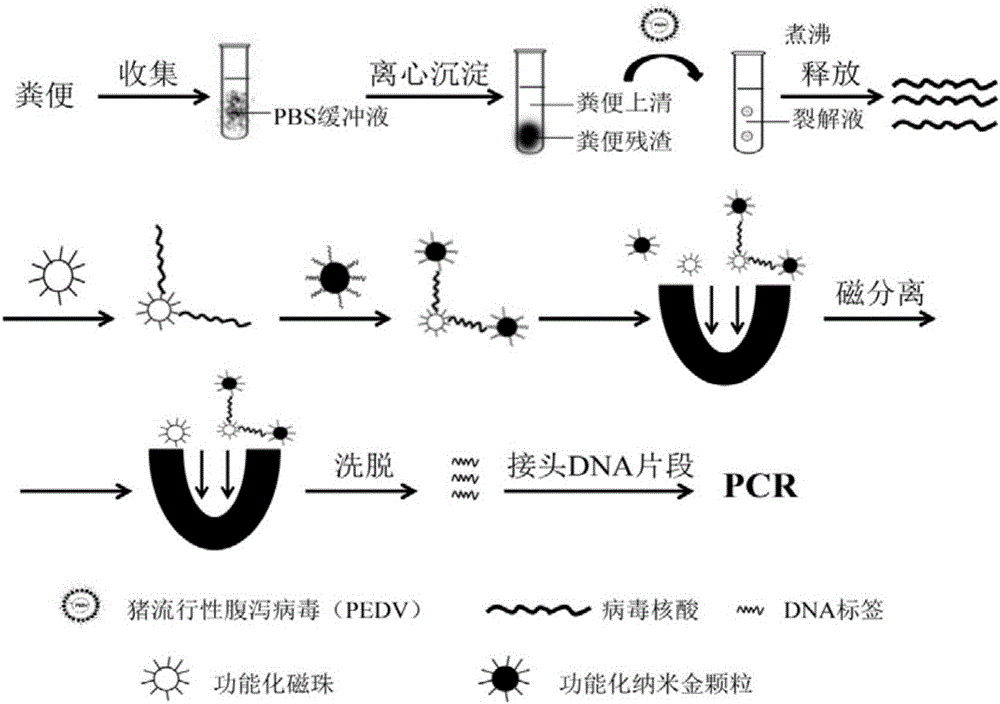
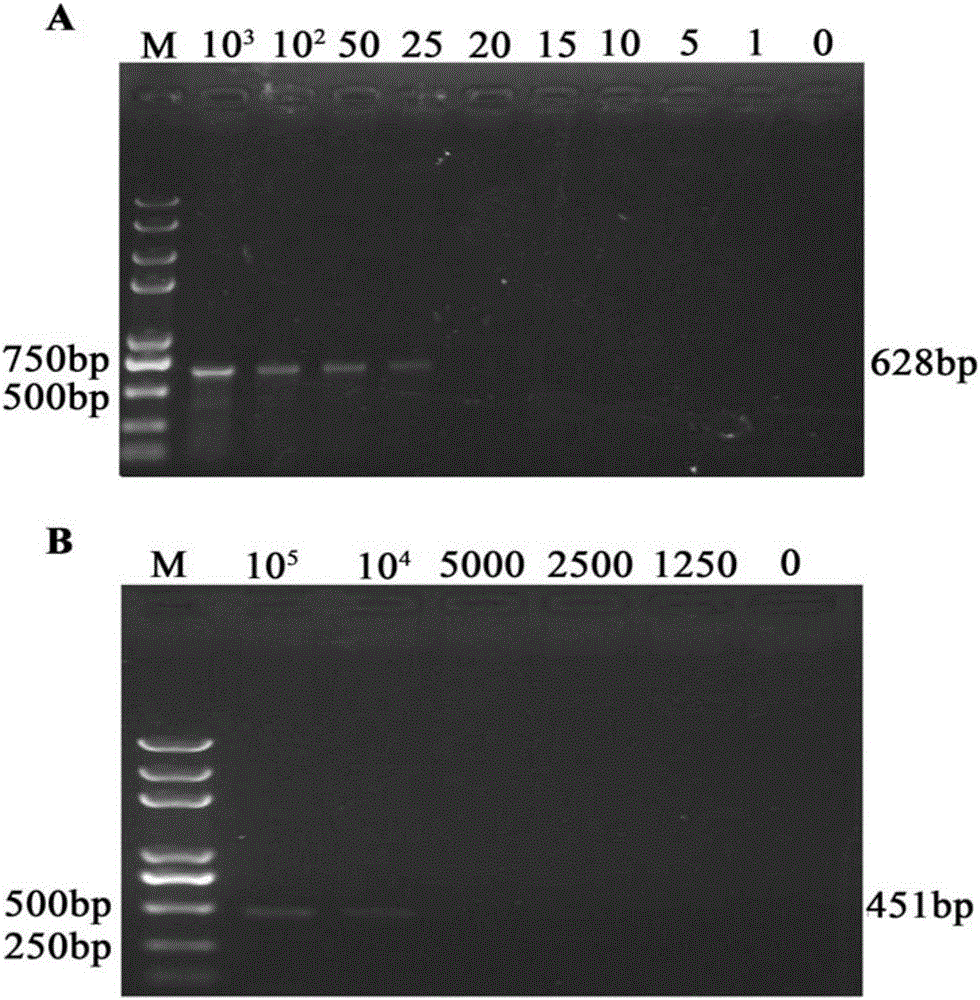
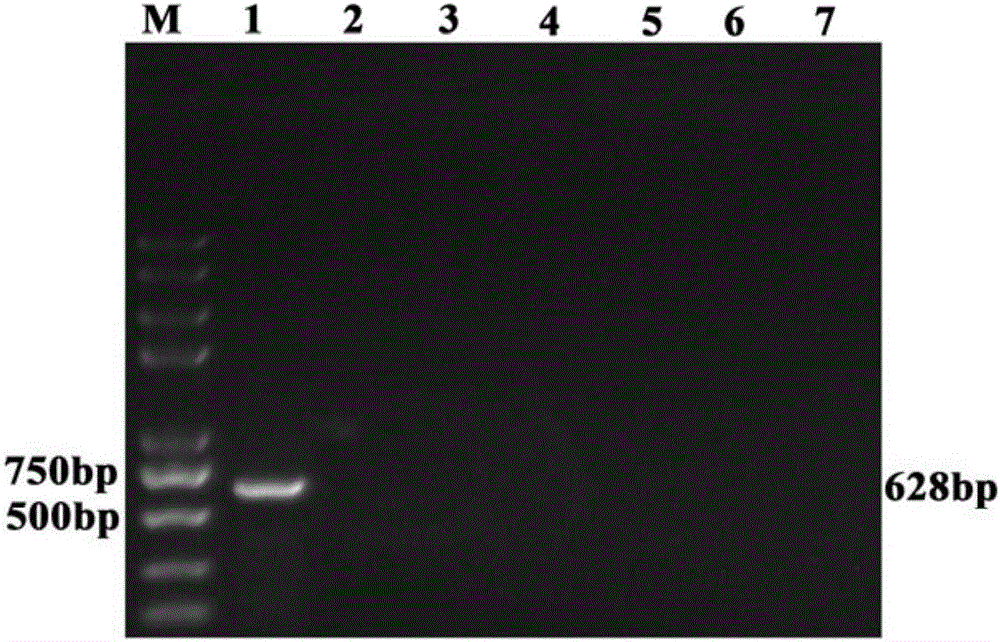
Method for rapidly detecting early-stage PEDV infection based on nanogold label amplification technology
InactiveCN105861746AStrong specificityIncreased sensitivityMicrobiological testing/measurementAnimal virusHybridization reaction
The invention provides a method for enriching viruses from faeces rapidly and efficiently, amplifying signals through gold nanoparticles crosslinked through specific DNA labels and then rapidly detecting early-state PEDV infection with a PCR. According to the method, an PRF1a gene located in a PEDV whole genome is adopted as a target sequence, specific nucleic acid probes are designed, second-segment probes which are high in specificity, great in virus enriching capability and easy to amplify and detect are screened out of the specific nucleic acid probes and optimized, and by means of functionalized magnetic particles and nanogold particles of the second-segment probes, a PCR detection technology based on nanoparticle PEDV specificity is established; meanwhile, by means of the technical method, the technical breakthrough of conducting a direct hybridization reaction between faeces sample lysate and the functionalized magnetic particles is achieved, the step of purifying a sample nucleic acid is completely omitted, specificity and sensitiveness are improved, meanwhile, the detection step is simplified, and detection time is shortened. By means of the method, time and labor are saved, cost is low, and in addition, the animal virus infection level can be prompted accurately.
Owner:NORTHWEST A & F UNIV
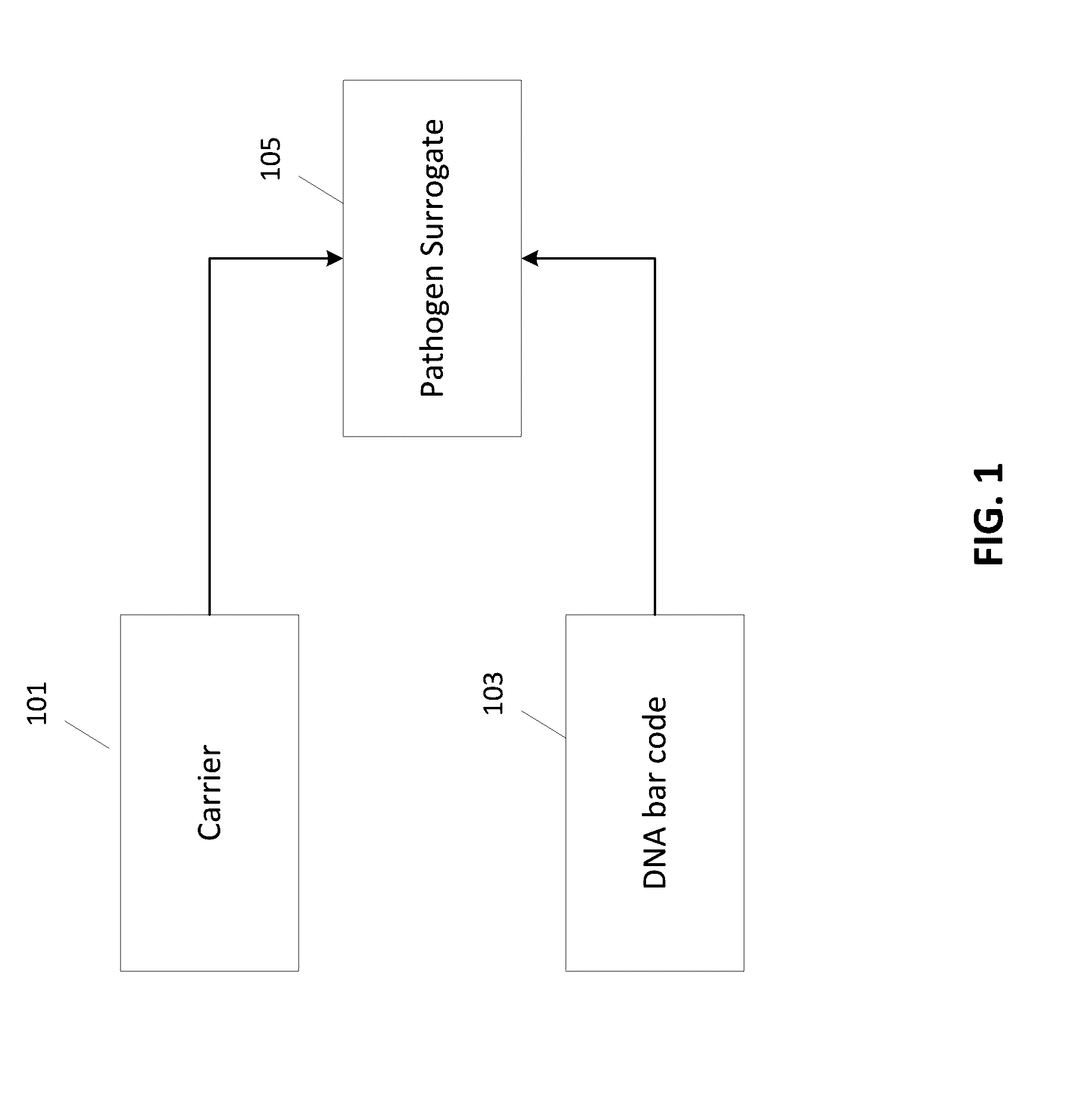
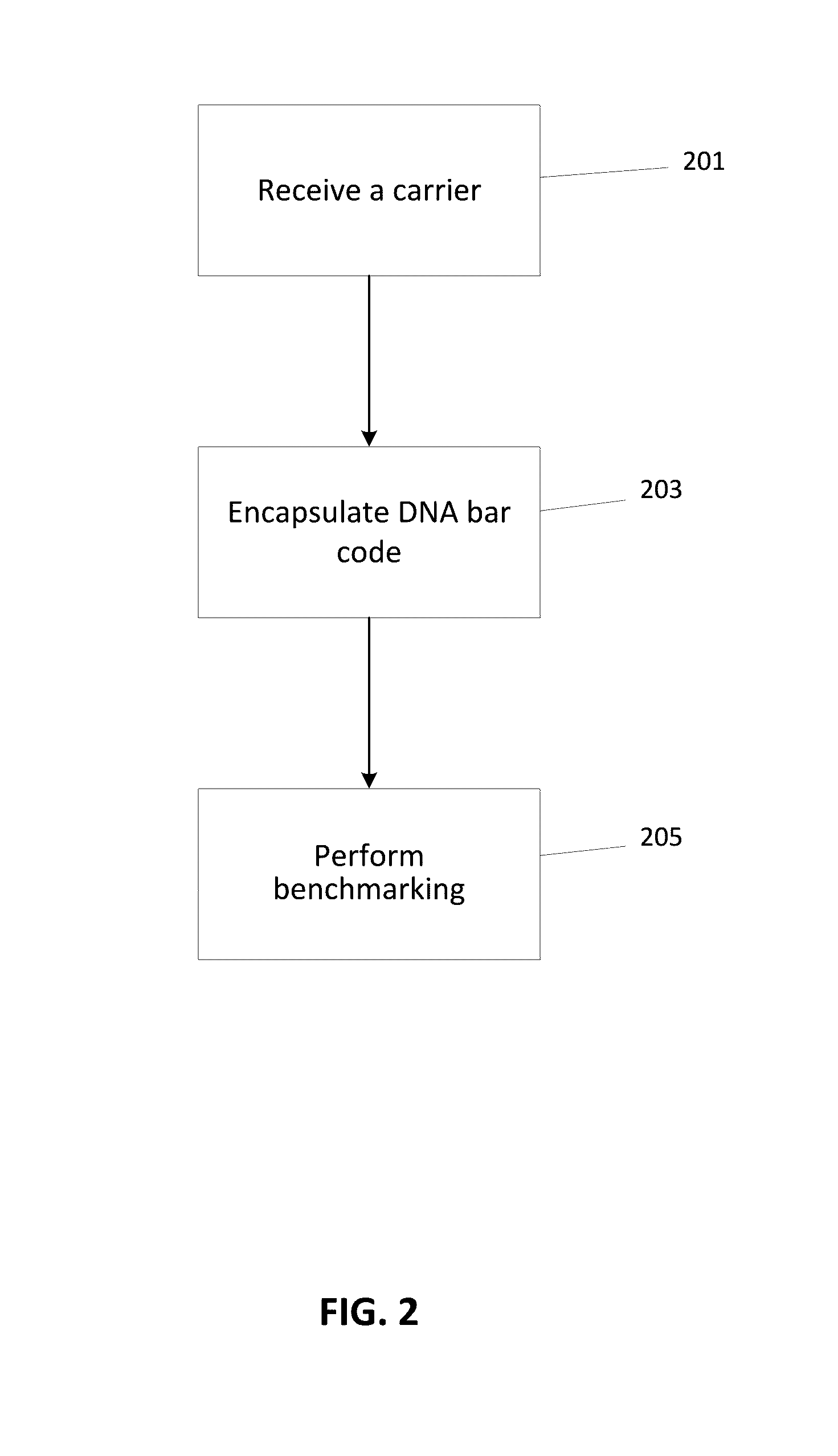
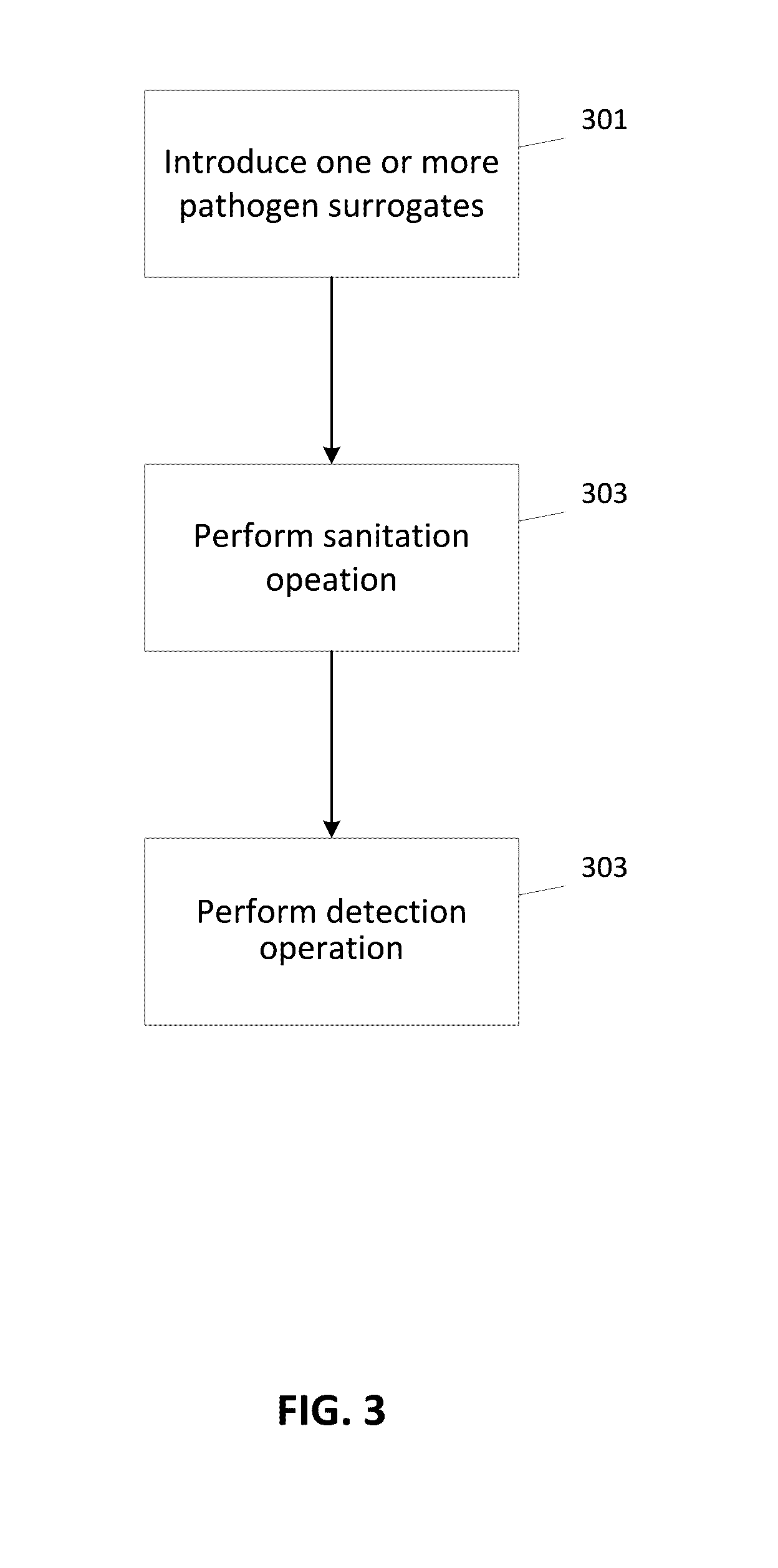
Pathogen Surrogates Based on Encapsulated Tagged DNA for Verification of Sanitation and Wash Water Systems for Fresh Produce
A pathogen surrogate, formed by a DNA tag or bar code and a carrier, is described for use in the validation and verification of sanitation, such as in food processing operations and for wash water systems for fresh produce. The carrier material is selected so that the pathogen surrogate mimics the behavior of a pathogen when subjected to a sanitation operation. One or more surrogates can be introduced in to an environment, which is then subjected to sanitation process, followed by a detection process using the DNA tag of the surrogate.
Owner:SAFETRACES INC
DNA markers for management of cancer
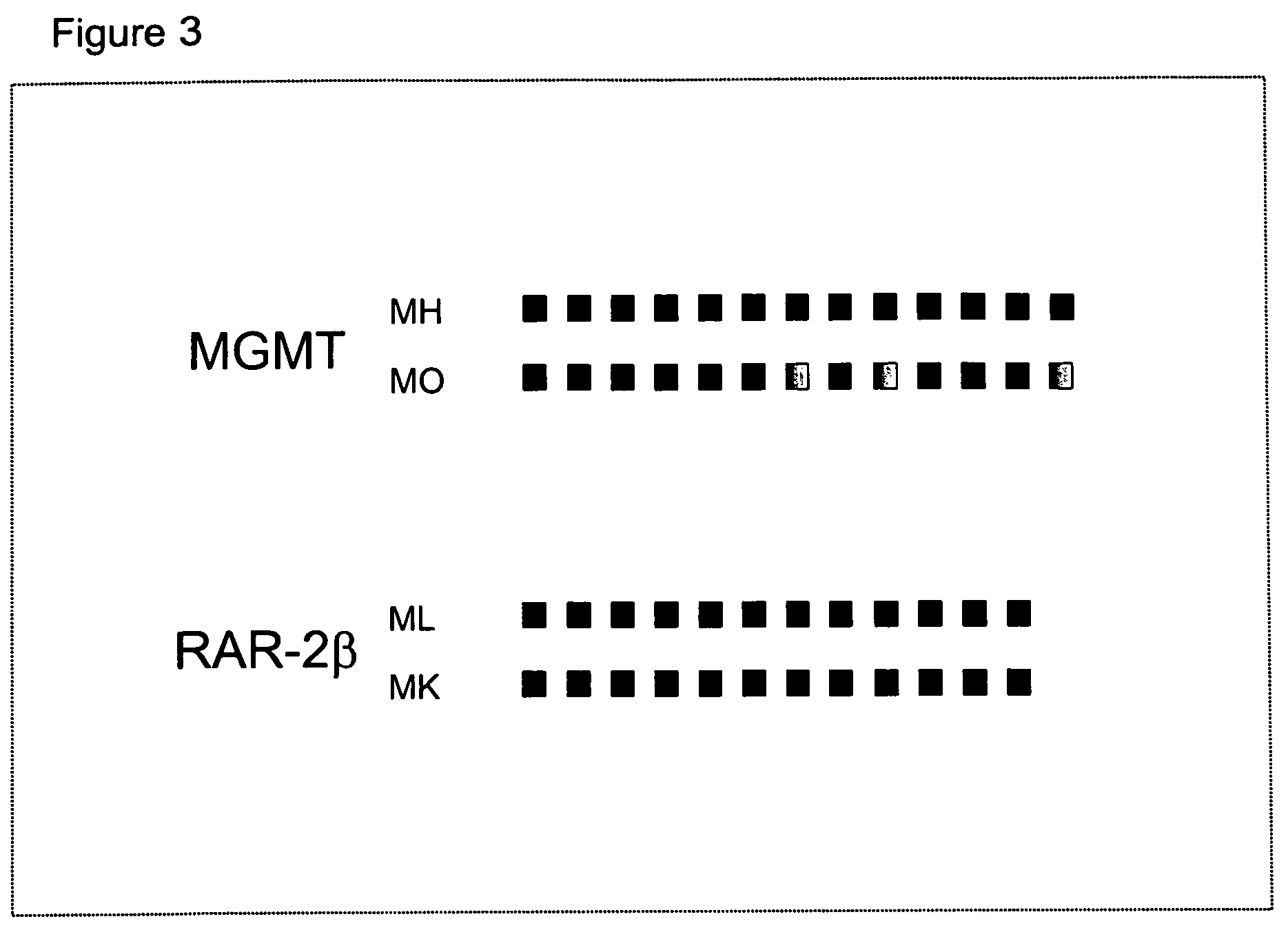
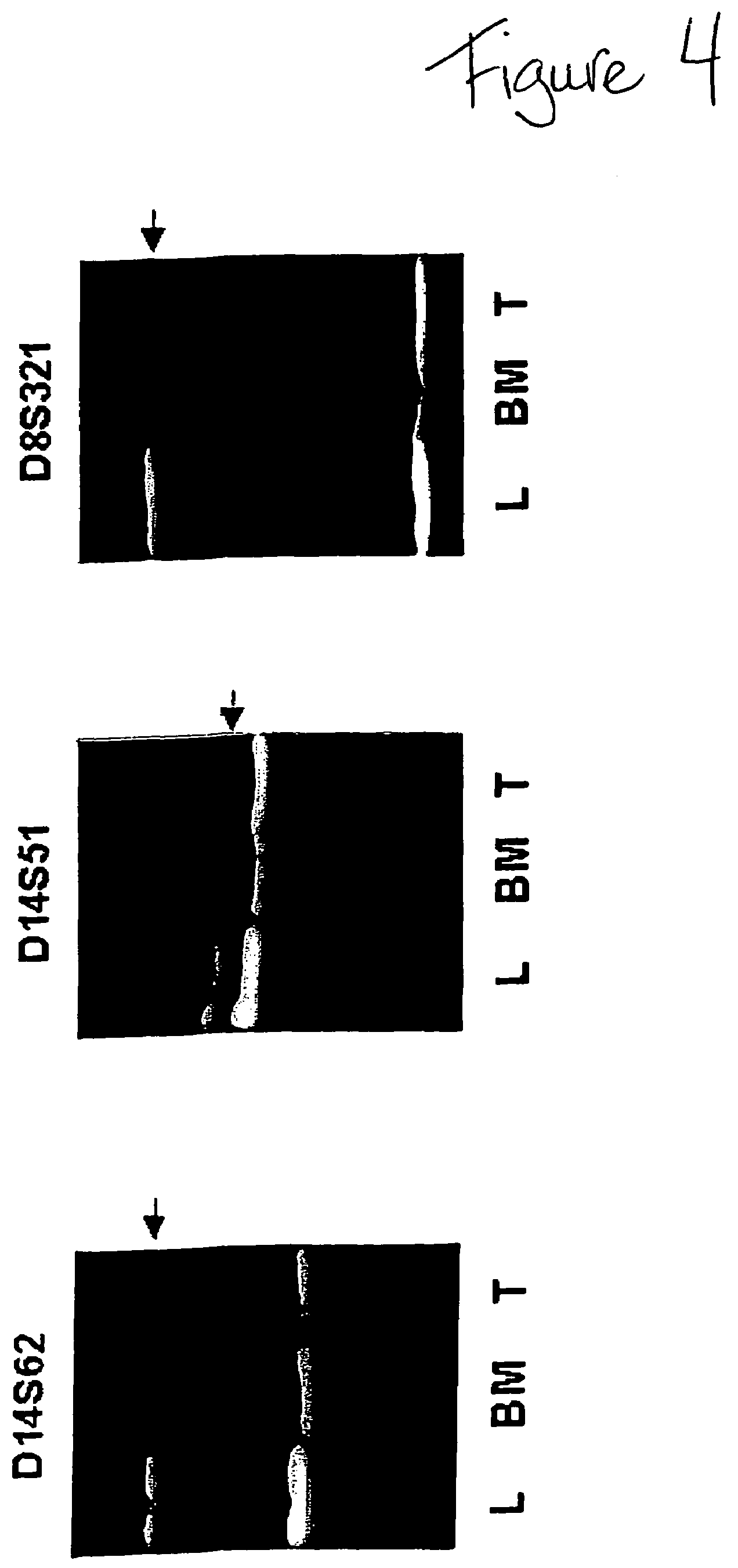
ActiveUS7718364B2Stage increaseMinimally invasiveSugar derivativesMicrobiological testing/measurementBlastomaSpecific chromosome
A method is provided for assessing allelic losses and hypermethylation of genes in CpG tumor promotor region on specific chromosomal regions in cancer patients, including melanoma, neuroblastoma breast, colorectal, and prostate cancer patients. The method relies on the evidence that free DNA and hypermethylation of genes in CpG tumor promotor region may be identified in the bone marrow, serum, plasma, and tumor tissue samples of cancer patients. Methods of melanoma, neuroblastoma, colorectal cancer, breast cancer and prostate cancer detection, staging, and prognosis are also provided.
Owner:JOHN WAYNE CANCER INST
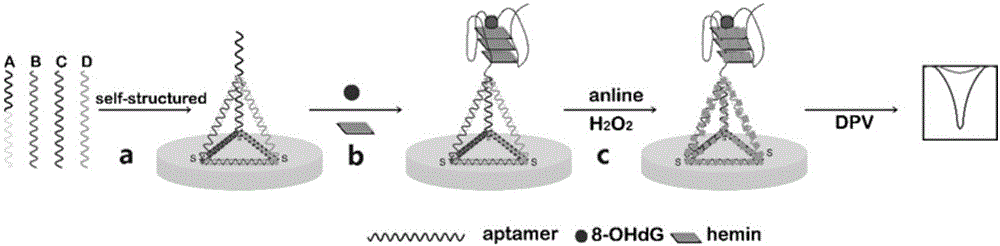
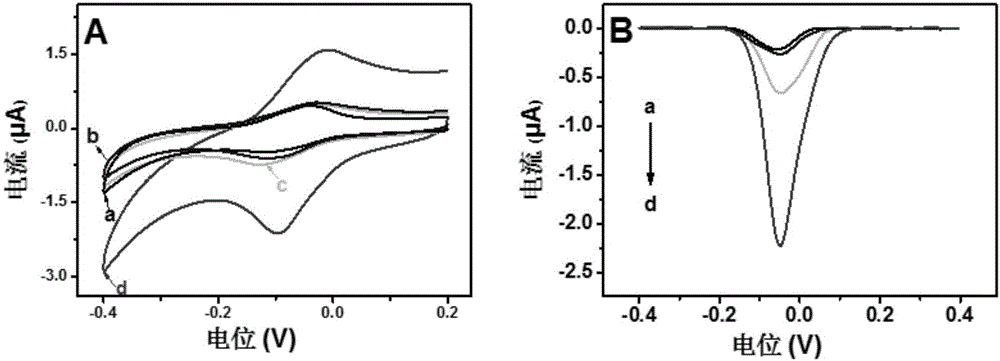
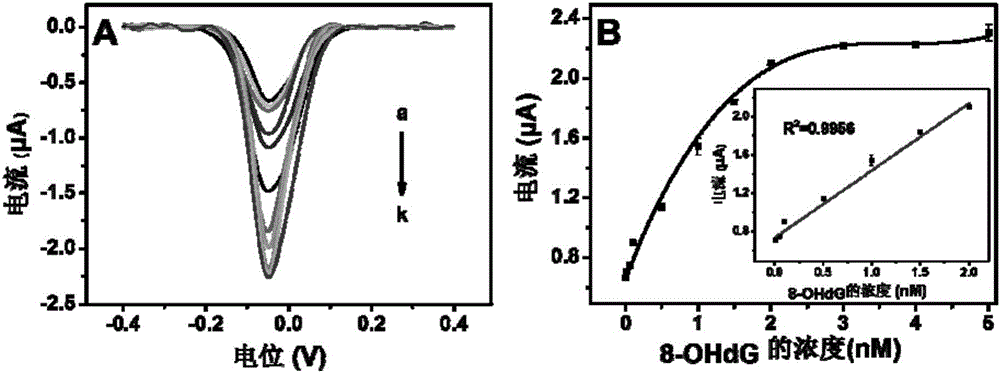
Method for quantitatively detecting activity of 8-OhdG (8-hydroxy-2'-deoxyguanosine) based on aniline deposited electrochemical sensing electrode
ActiveCN106442659AGuaranteed OrientationGuaranteed distanceMaterial electrochemical variablesAptamerPower flow
The invention discloses a method for quantitatively detecting the activity of 8-OhdG (8-hydroxy-2'-deoxyguanosine) based on an aniline deposited electrochemical sensing electrode. The method comprises the following steps of preparing a DNA (Deoxyribonucleic Acid) tetrahedral structure with sulfhydryl; modifying, on a gold electrode, the DNA tetrahedral structure with the sulfhydryl to obtain an electrode of which the top end is connected with an 8-OhdG aptamer and which has the DNA tetrahedral structure with the sulfhydryl; forming a G-quadplex electrode; forming a polyaniline deposited electrochemical sensing electrode; detecting a generated current signal of polyaniline by utilizing an electrochemical method, so as to detect the activity of the 8-OhdG. By using the method for quantitatively detecting the activity of the 8-OhdG based on the aniline deposited electrochemical sensing electrode, the detection sensitivity on the 8-OhdG is greatly improved. In compassion with a conventional electrochemical detection method using a reduction peak of the 8-OhdG as a signal, the detection limit is decreased by two orders of magnitude. By using the method for quantitatively detecting the activity of the 8-OhdG based on the aniline deposited electrochemical sensing electrode, the preparation of a complicated material and a DNA labeling probe is not needed; the defects that the preparation of the material and the DNA labeling probe causes that the detection cost is high, the operation is fussy and the reproducibility is poor can be avoided. The method for quantitatively detecting the activity of the 8-OhdG based on the aniline deposited electrochemical sensing electrode has the advantages of being low in cost, being quick, simple and convenient, and being high in sensitivity.
Owner:SOUTHEAST UNIV
Environmental microorganism detection method and system
InactiveCN101429559AMicrobiological testing/measurementSpecial data processing applicationsMicroorganismHigh flux
The invention is suitable for the field of bioengineering and provides a method and a system for detecting environmental microorganisms. The method comprises the following steps: DNA extracted from an environmental sample is sequenced by the high-flux sequencing technology, and DNA label sequence is obtained; vector pollution in the DNA label sequence is removed; and the DNA label sequence is compared with the known sequence in a known database, and the class of the DNA label sequence is determined according to the comparison result. The embodiment of the method can detect possible microbial species and possible varieties of microbial species in the environmental sample.
Owner:SHENZHEN HUADA GENE INST +1
Noninvasive biopsy virus detection method based on high throughput gene sequencing and tagged connector
ActiveCN105567681AImprove throughputHigh speedMicrobiological testing/measurementLibrary creationDiseaseVirus detection
The invention provides a noninvasive biopsy virus detection method based on high throughput gene sequencing. The method can carry out DNA sequencing and RNA sequence in parallel and thus precisely determines the biological composition and structure of viruses in patients. The method can maximally avoid the interference of nucleic acid of host cells, effectively reduce the cost of sequencing, and is capable of detecting multiple unknown crossed viruses at the same time. 80 DNA tagged connectors are designed, related reagents are assembled to form a library so as to construct a kit, and if the throughput allows, the kit can perform sequencing on 80 samples in parallel. The experiment design is strict, the automation degree is high, the virus identification can be finished in batches within 24 hours, the requirements of scientific research and clinical detection can be met in different levels, and references are provided for clinical diagnosis and prescription of diseases related with virus infection.
Owner:GUANGZHOU SAGENE BIOTECH
DNA label sequence, sequencing library construction method and kit
ActiveCN105039322AStrong specificityNo non-specific bindingNucleotide librariesMicrobiological testing/measurementA-DNARepeatability
The invention relates to a DNA label and PCR primer pairs with the 5'-end connected with the DNA label, and provides a sequencing library construction method adopting the DNA label and the PCR primer pairs, and a kit composed of the DNA label and the PCR primer pairs. The DNA label and amplimers form a detection product with optimized and balanced specificity, sensitivity and repeatability, the detection product can be used to detect 1-20 different sources of a sample once, and can accurately distinguish the base sequence of various sample sources. The coincidence rate of high flux sequencing and a sequencing method reaches up to 100%.
Owner:SUREXAM BIO TECH
Detection method for Apostichopus japonicas AjE101 micro-satellite DNA label
InactiveCN102140522AGood polymorphismImprove stabilityMicrobiological testing/measurementGerm plasmGenetic diversity
The invention belongs to the field of molecular biology DNA labeling technology and application, and in particular relates to Apostichopus japonicas express sequence tag EST micro-satellite label screening development and application. The invention provides Apostichopus japonicas AjE101 micro-satellite specific DNA primers, a polymerase chain reaction (PCR) reaction system by utilizing the primers, and a detection method for an Apostichopus japonicas AjE101 micro-satellite DNA label. The detection method comprises the following steps of: extracting genome of Litopenaeus vannamei Boone; designing the specific primers at two ends of an Apostichopus japonicas AjE101 micro-satellite DNA core sequence; and performing PCR amplification on genome DNA of different groups of the Litopenaeus vannamei Boone or individuals in the group by using the primers, analyzing products, and determining the genome of each individual so as to obtain an Apostichopus japonicas polymorphism genetic variation map. The invention is mainly applied to Apostichopus japonicas germ plasm resource and genetic diversity analysis, cular population genetics, construction of genetic maps and research of functional genes.
Owner:SOUTH CHINA SEA INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Method for cloning rice auxin induced protein gene
InactiveCN101492671AHigh homologySimple implementation stepsFermentationPlant genotype modificationMutantDNA Intercalation
The invention relates to a method for using rice auxin to induce protein genes to be cloned, which is characterized in that the implementation steps comprise: (1) the preparation of a rice transformation receptor; (2) the genetic transformation of the rice; (3) the screening of kanamycin-resistant callus tissue and the regeneration of plants; (4) the screening of the mutant of a T-DNA inserted progenies; (5) Tail-PCR; (6) the comparison and analysis of sequences on the Internet. In the invention, a rice mutant with a short plant height is obtained when carrying out rice functional genome research by using T-DNA label method; the lateral neighboring sequences of the mutant are researched by using TAIL-PCR technology; meanwhile, the position where the mutant T-DNA inserts the rice genome is arranged on the No. 4 chromosome of the rice by the comparison on the databases of NCBI and TIGR on the Internet; moreover, the rice BAC clone (OSJNBa0084K01) of the position is found out. The T-DNA is inserted between the two genes of the clone by analyzing the clone. Known functional genes with a very high homology with BAC cloning code amino acid sequence are forecasted by the sequence comparison on the Internet.
Owner:TIANJIN AGRICULTURE COLLEGE
Primer, method and kit for constructing gene mutation sequencing library
ActiveCN104946639AGood amplification effectMultiplex PCR amplification effect is goodMicrobiological testing/measurementLibrary creationMicrobiologyA-DNA
The invention relates to a PCR primer involving 47 genes and a tag PCR primer. The tag PCR primer is composed of the PCR primer and a DNA tag sequence connected to the 5' end of at least one primer in the PCR primer pair, wherein the DNA tag sequence is selected from SEQ ID NO: 107 to SEQ ID NO: 126. The invention further relates to a method and a kit for constructing a sequencing library by applying the DNA tag of the tag PCR primer. The coincidence rates of the detection method provided by the invention and the sequencing method is up to 100%, the provided amplification primer and the DNA tag form a detection product with a specificity, a sensitivity and a repeatability which are optimized and balanced, somatic mutation and mononucleotide polymorphism can be detected in parallel, and a plurality of base sequences of 1-20 sample sources can be accurately differentiated by one-time detection.
Owner:广州益善医学检验所有限公司
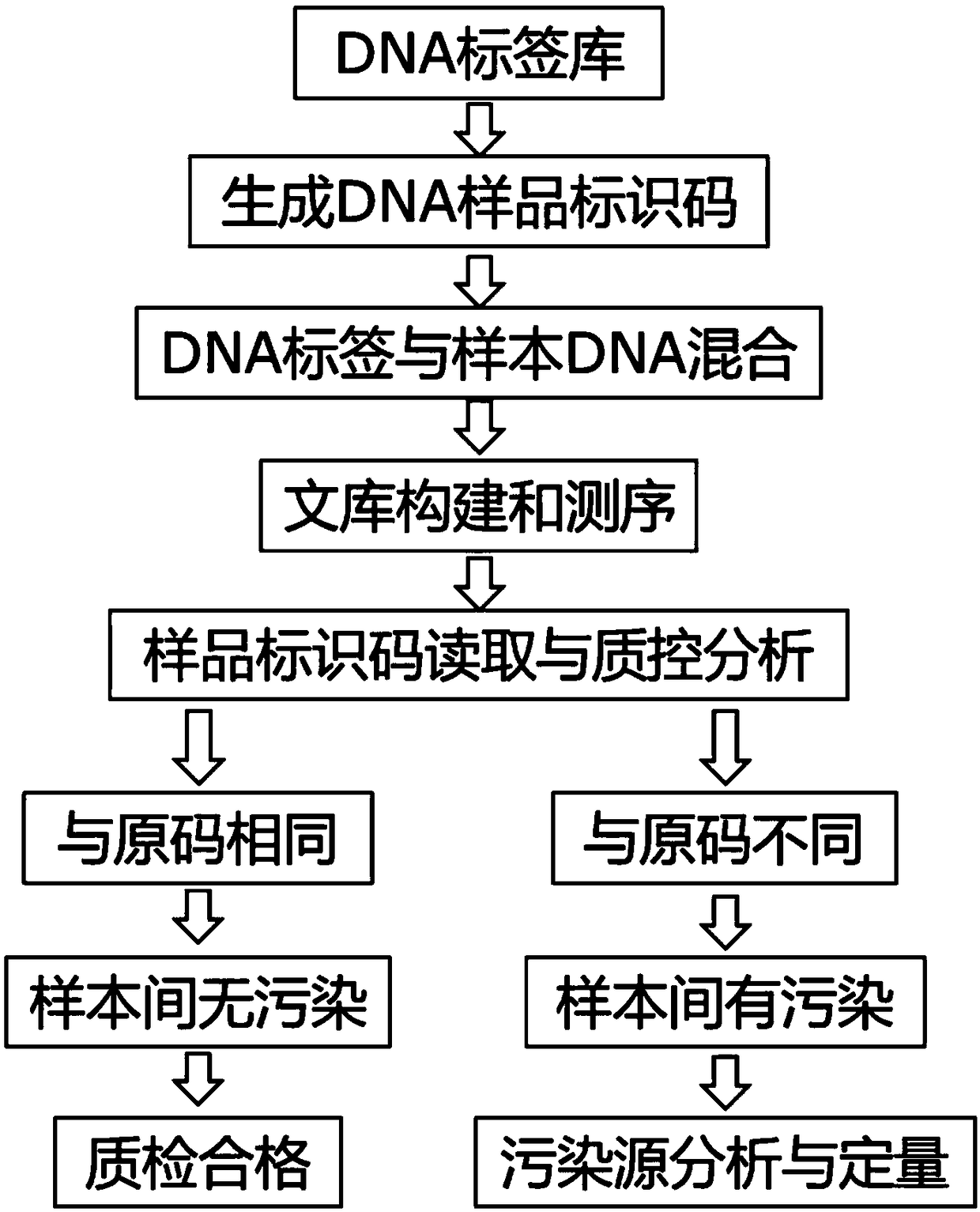
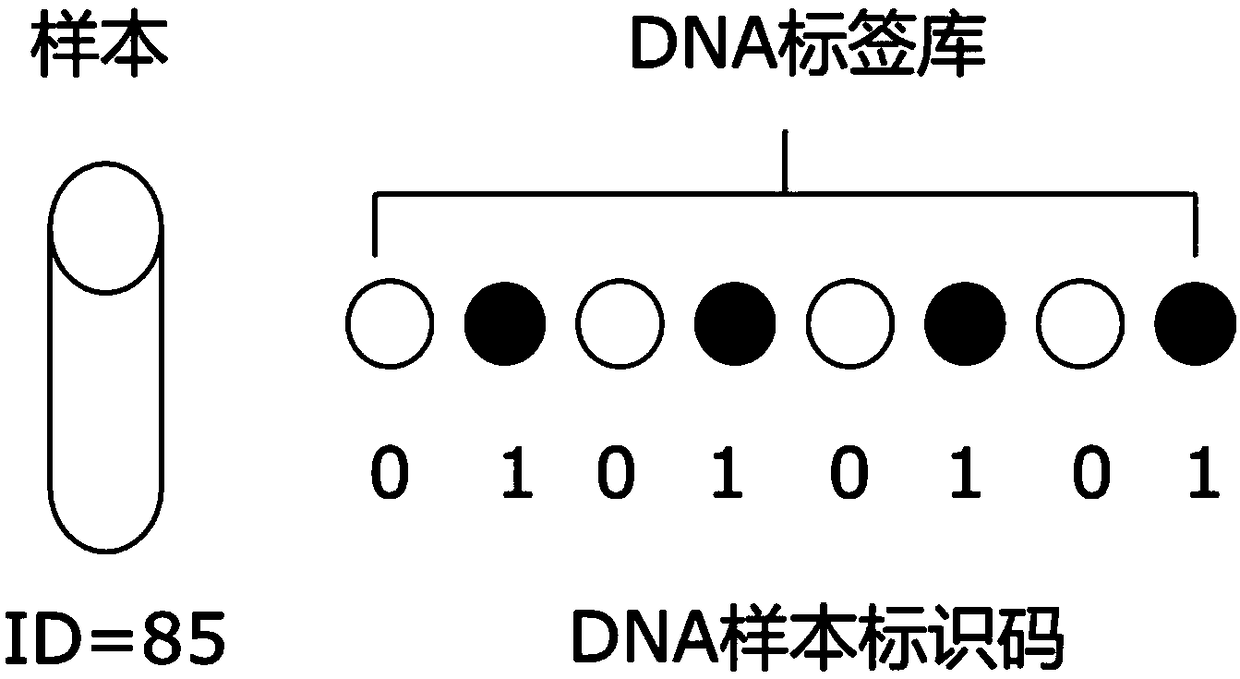
Identification method for sequencing sample and application thereof
ActiveCN108932401AEasy to readEasy to analyzeMicrobiological testing/measurementSpecial data processing applicationsComputational biologyDna labeling
The invention provides an identification method for a sequencing sample and an application thereof. The method comprises the following steps: (1) constructing an injective relationship between a sequencing sample library and a DNA tag library; (2) according to the injective relationship described in the step (1), using a DNA tag in the DNA tag library to identify a sequencing sample. The method determines a DNA tag combination uniquely corresponding to the sequencing sample by constructing the injective relationship between the sequencing sample library and the DNA tag library, and different sequencing samples are corresponding to different DNA tag combinations, thereby realizing specificity identification of the sequencing samples by the DNA tags.
Owner:江西海普洛斯生物科技有限公司
DNA tags, PCR primer and application thereof
ActiveCN105316320AHigh resolutionAvoid troubleBioreactor/fermenter combinationsBiological substance pretreatmentsNucleotideGenotype
The invention discloses DNA tags, a PCR primer and application thereof. The DNA tags of one group are chosen from nucleotides shown as in SEQ ID NO:1-95 and can be used for building a nucleic acid sequencing library to accurately differentiate the nucleic acid sequencing library. The DNA tags and the PCR primer are utilized to build a tagged PCR primer, and by utilizing the tagged PCR primer, STR detection of at most 95 DNA samples can be realized at one step according to a method for determining preset STR gene locus genotype of various DNA samples.
Owner:TIANJIN MEDICAL LAB BGI +1
Optical mapping of genomic DNA
InactiveUS20130130255A1High densityHigh precisionBioreactor/fermenter combinationsBiological substance pretreatmentsGenomic DNAOptical mapping
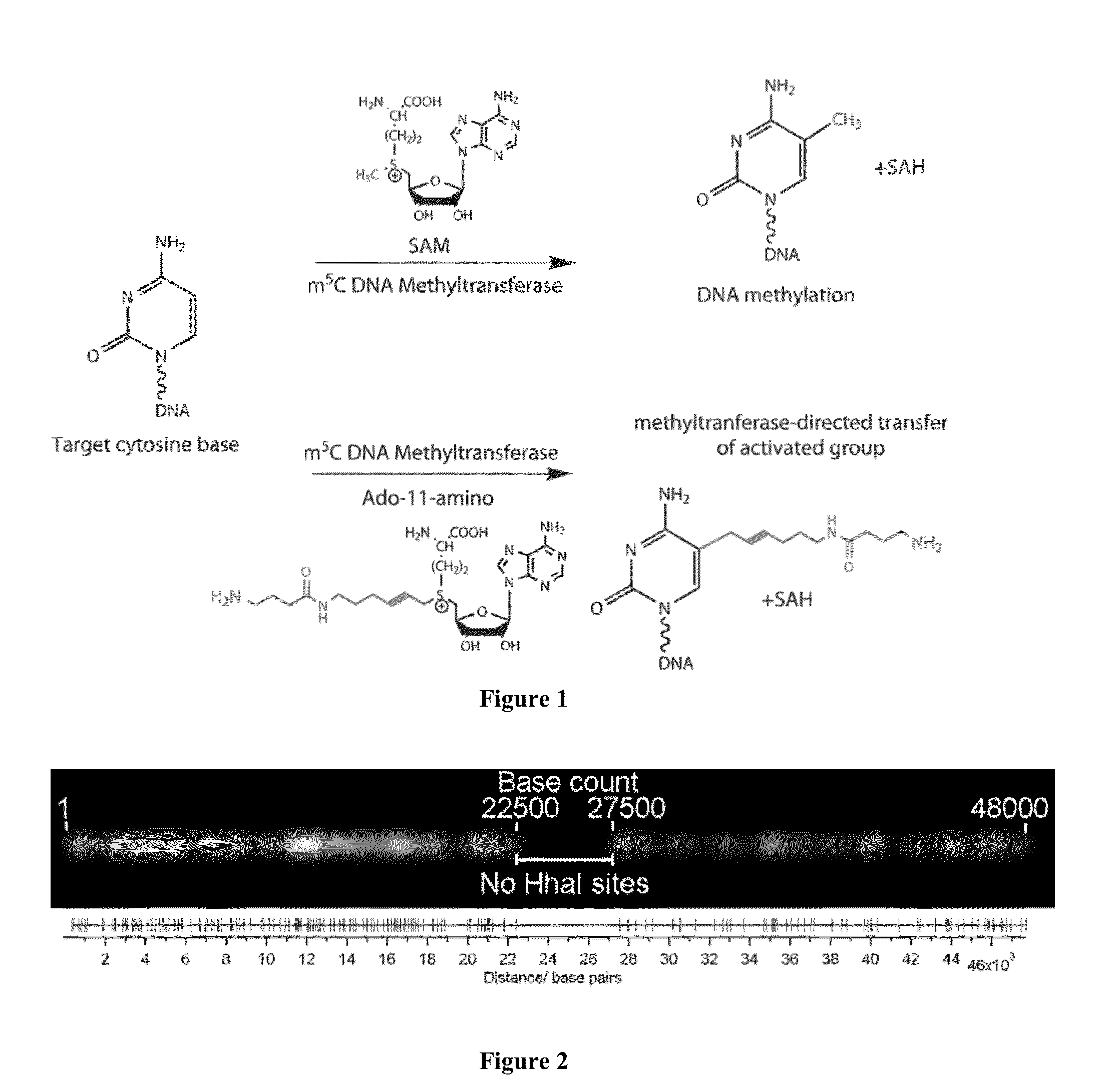
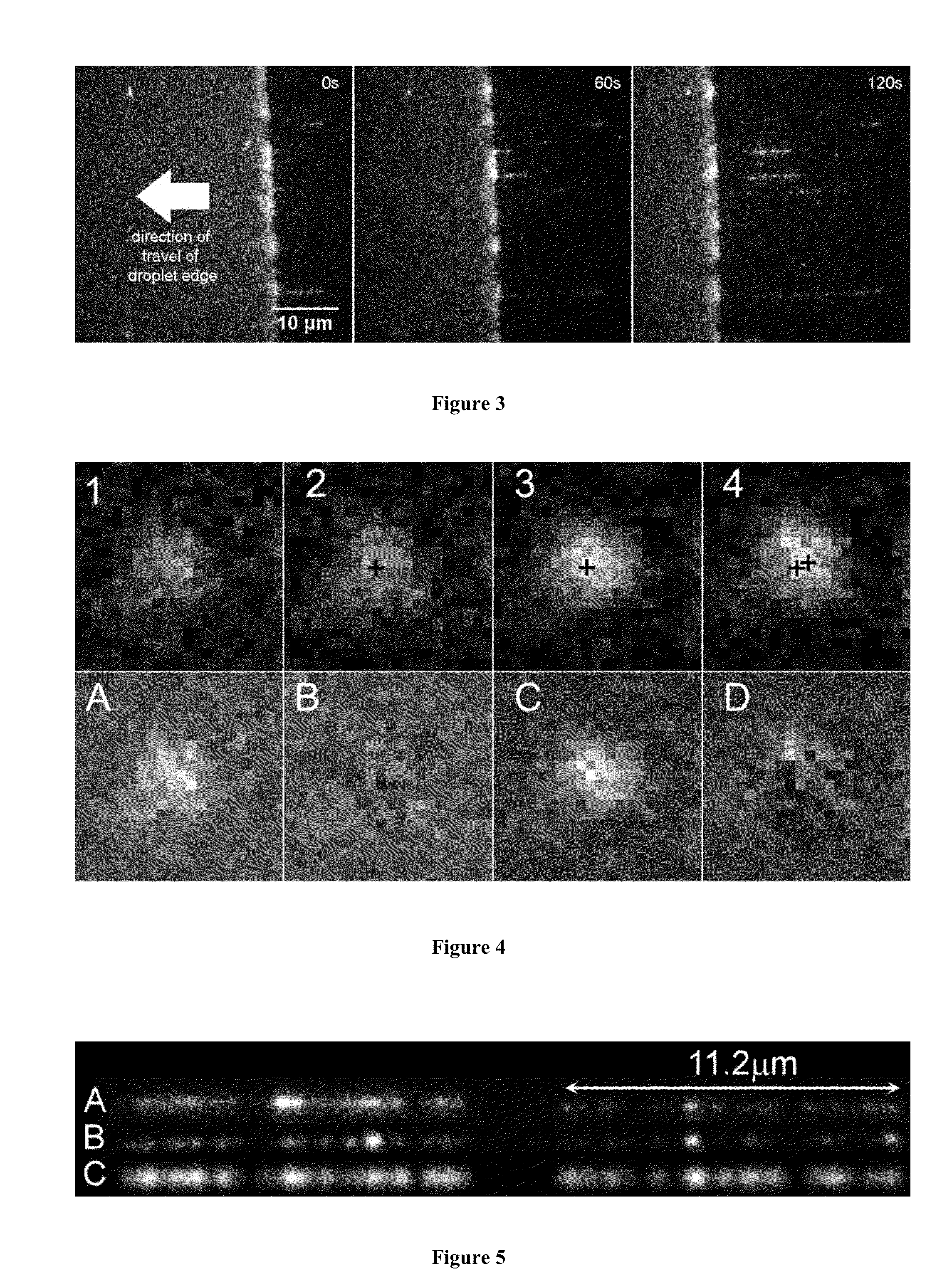
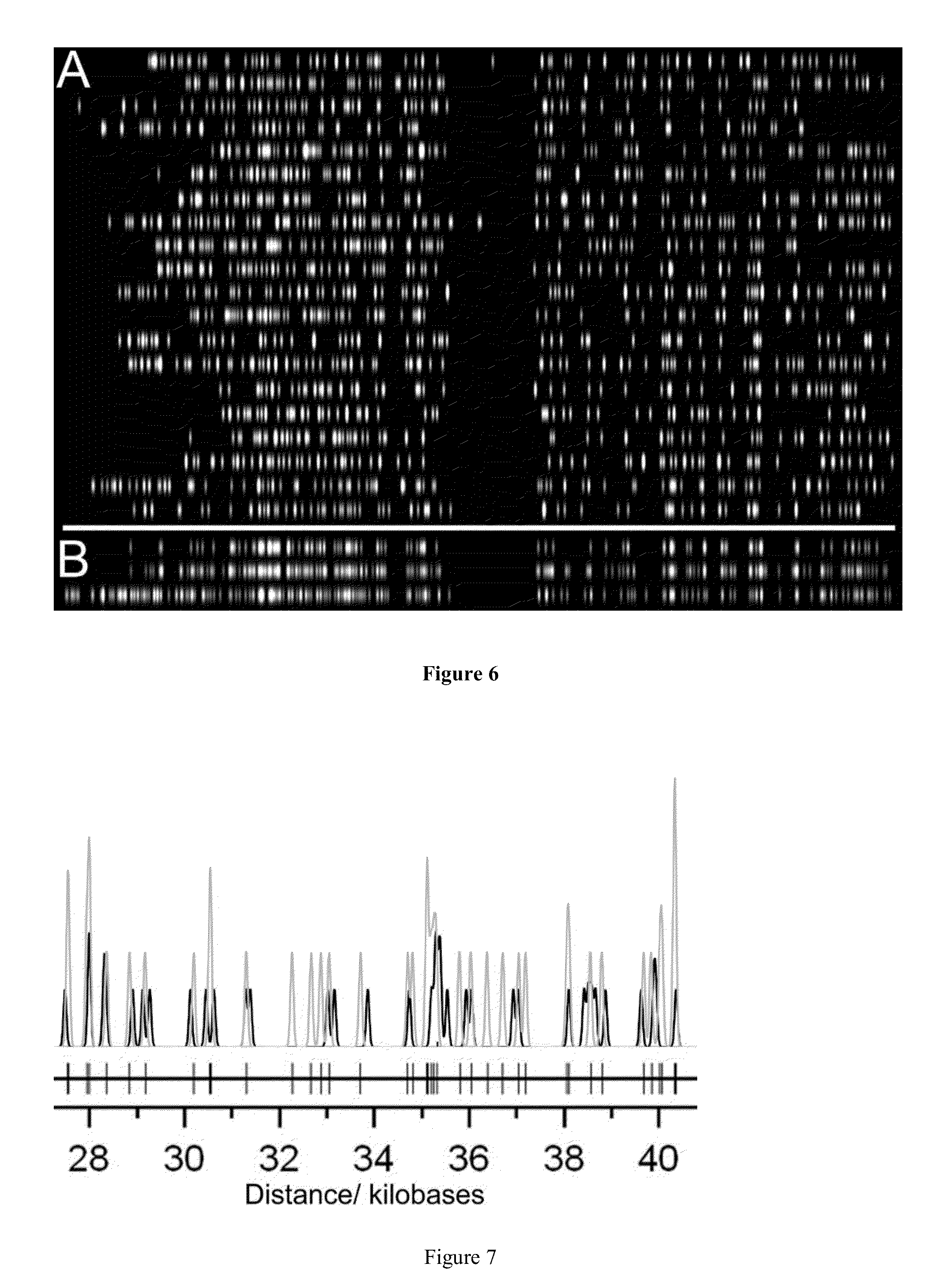
A method for single-molecule optical DNA profiling using an exceptionally dense, yet sequence-specific coverage of DNA with a fluorescent probe, using a DNA methyltransferase enzyme to direct the DNA labeling, followed by molecular combing of the DNA onto a polymer-coated surface and subsequent sub-diffraction limit localization of the fluorophores. The result is a ‘DNA fluorocode’; a simple description of the DNA sequence, with a maximum achievable resolution of less than 20 bases, which can be read and analyzed like a barcode. The method generates a fluorocode for genomic DNA from the lambda bacteriophage using a DNA methyltransferase to direct fluorescent labels to four-base sequences reading 5′-GCGC-3′. A consensus fluorocode is constructed that allows the study of the DNA sequence at the level of an individual labeling site and is generated from a handful of molecules and entirely independently of any reference sequence.
Owner:KATHOLIEKE UNIV LEUVEN
DNA label, PCR primer and application thereof
ActiveCN105296471AHigh sequencing throughputReliable test resultsBioreactor/fermenter combinationsBiological substance pretreatmentsNucleotideNucleic acid sequencing
The invention discloses a DNA label, a PCR primer and application thereof. The DNA label is chosen from nucleotides shown as SEQ ID NO:1-95 and can be used for building a nucleic acid sequencing library to accurately differentiate the nucleic acid sequencing library. The DNA label and the PCR primer are utilized to structure a label PCR primer, and deafness gene mutation detection of 95 DNA samples can be realized at most in one time by utilizing the label PCR primer according to a method for determining whether deafness genes of various DNA samples are mutational or not.
Owner:TIANJIN MEDICAL LAB BGI +1
Method of organism identification by DNA tag
InactiveCN101379188ADoes not cost laborImport indirectMicrobiological testing/measurementRecombinant DNA-technologyA-DNAOrganism
A novel method of organism identification that can replace the conventional organism identification method by DNA requiring a large amount of labor. There has been developed a method of organism identification, namely, a method of identifying whether or not being a specified organism or a descendant thereof, characterized in that whether or not an identification object organism is a specified organism or a descendant thereof is judged by introducing in advance a DNA tag in cells of the specified organism or organism living parasitically or symbiotically therewith and finding whether the DNA tag is detected or not from the DNA extracted from the identification object organism or organism living parasitically or symbiotically therewith.
Owner:NIPPON SOFTWARE MANAGEMENT +2
Label and preparation method and applications thereof
ActiveCN108165620AImprove stabilityImprove label recognition rateMicrobiological testing/measurementLibrary creationA-DNABioinformatics
The invention discloses a label preparation method, a label, a label connector, and a gene sequencing method based on a DNA label library. The provided label preparation method can rapidly obtain a label sequence. The label screening and testing period is short. 384 labels and label connectors obtained by the method can be applied to DNA label library building and gene sequencing, sequencing on 384 samples can be carried out parallelly, the stability is good, the label recognition rate and the accuracy are high, and the label is especially suitable for an Ion torrent platform.
Owner:CAPITALBIO GENOMICS
Method for rapidly and accurately breeding sweet and glutinous double recessive corn selfing line
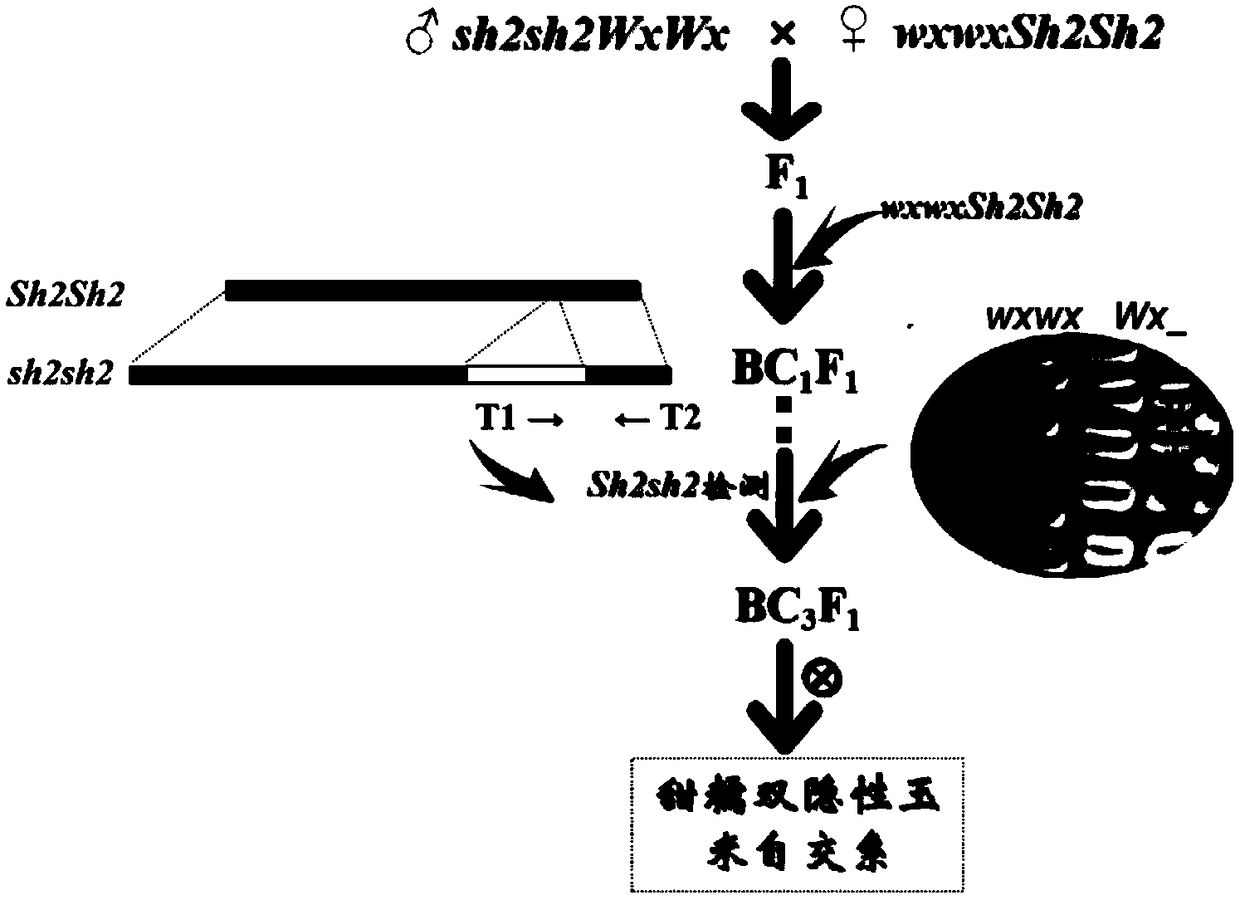
PendingCN108812298AImprove breeding efficiencyAccurate and efficient screeningMicrobiological testing/measurementPlant genotype modificationBiotechnologyGermplasm
The invention discloses a method for rapidly and accurately breeding a sweet and glutinous double recessive corn selfing line. The method comprises following steps: taking a sh2sh2 type super-sweet corn as a donor parent and a wxwx corn that is in accord with the breeding target as a receptor parent to establish BC1F1; dyeing BC1F1 seeds by an iodine staining method, screening wxwx germplasm namely seeds with a light color; subjecting wxwx germplasm to DNA mark detection to screen out germplasm with SH2sh2, for each backcrossing generation, carrying out iodine staining and DNA labeling to obtain target seeds, backcrossing to BC3F1, and carrying out selfing to obtain BC3F2, wherein the wrinkled seeds are the sweet and glutinous double recessive corn selfing line. The method has the advantages that specific primers (T1, T2) are used to carry out detection so as to accurately and efficiently screen out individuals containing a recessive sh2 gene, and through the directional selection of backcrossing generation, the breeding efficiency is obviously improved.
Owner:HEBEI AGRICULTURAL UNIV.
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com