DNA labels, PCR primers and application thereof
A label and mixture technology, which is applied in DNA label, PCR primer and its application field, can solve the problems that the deafness gene detection technology needs to be improved.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] According to the method of determining whether there is a mutation in the deafness gene of multiple DNA samples according to the present invention, the deafness gene detection is carried out, specifically as follows:
[0079] (1) Primer design
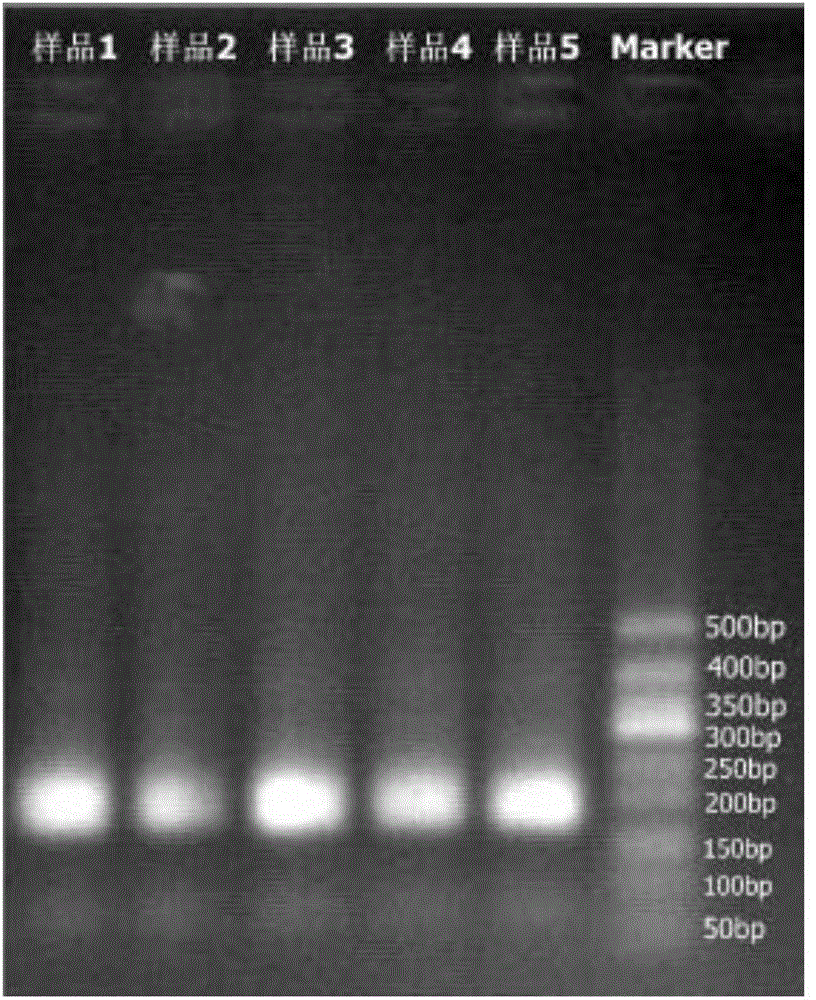
[0080] According to the CDS region and 12S rRNA of GJB2, GJB3 and SLC26A4 genes, the primer design software was used to design the amplification primers of the above gene loci. The length of the amplification primers was about 20 bases. Use software to design and evaluate the DNA index (index) sequence. The index sequence contains 7 bases. When designing the index sequence, it should avoid sequences with high similarity to the sequencing primers and the tagged PCR amplification primers to form a hairpin structure or dimerization. Body and other secondary structures. Refer to Table 1 for the amplification primers designed by the present invention for the above-mentioned GJB2 and GJB3 gene CDS regions, SLC26A4 gene No. 2-21 exon ...
Embodiment 2
[0164] According to the method of determining whether there is a mutation in the deafness gene of multiple DNA samples according to the present invention, the deafness gene detection is carried out, specifically as follows:
[0165] (1) For samples 1-5 in Example 1, repeat the experimental operations of primer design, PCR amplification and amplified product mixing, end repair, adapter ligation, and library detection in Example 1, and also obtain the samples in Example 1. Library samples of known concentration, for later use.
[0166] (2) Preparation of Ion PGM TM Sequencing template
[0167] ①One Touch TM 2
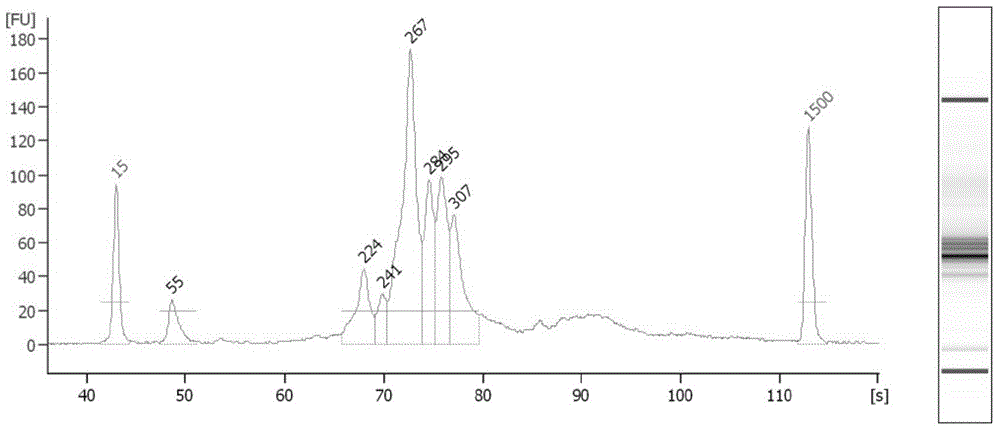
[0168] Detect the library concentration according to the Agilent Bioanalyzer2100, dilute the library to 80pM, prepare the OneTouch 2 reaction mixture according to Table 8, transfer the adapter and load it into the initialized OneTouch TM 2. See OneTouch for details on the operation process TM manual.
[0169] Amplification solution, respectively, 1000 μL of amplifi...
Embodiment 3
[0181] According to the method of determining whether there is a mutation in the deafness gene of multiple DNA samples according to the present invention, the deafness gene detection is carried out, specifically as follows:
[0182] (1) Primer design: According to the selected 20 deafness gene mutation sites to be detected and / or typed, they are: 35delG, 167delT, 176-191del16, 299_300delAT and 235delC of the GJB2 gene, 538C>T of the GJB3 gene 547G>A, 281C>T, 589G>A, IVS7-2A>G, 1174A>T, 1226G>A, 1229C>T, 1975G>C, 2027T>A, 2162C>T, 2168A>G and IVS15+5G>A, 1494C>T and 1555A>G of 12S rRNA gene. The amplification primers designed by the present invention for the 20 mutation sites of the above four deafness genes are selected from the primers in Example 1, see Table 15.
[0183] Table 15
[0184]
[0185]
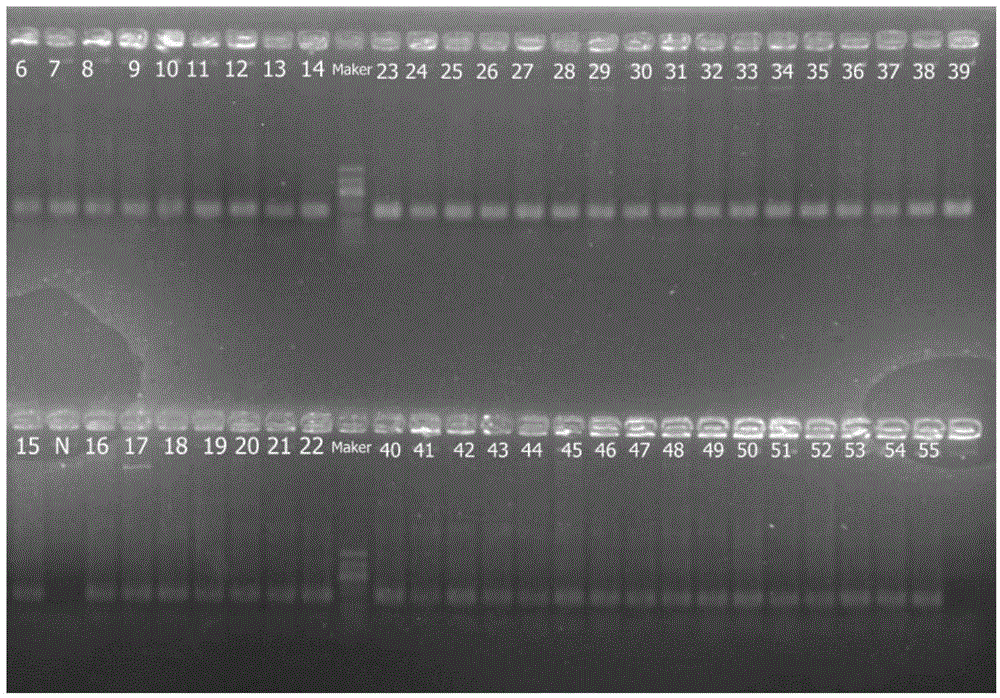
[0186] (2) In addition to the different samples tested, subsequent experimental steps (ie: PCR amplification and amplification product mixing, end repair, adapter ligatio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com