Label and preparation method and applications thereof
A tag and tag pool technology, applied in biochemical equipment and methods, DNA preparation, chemical library, etc., can solve the problems of high sequencing error rate, tag sequence change, misjudgment as another tag, etc., and achieve a high tag recognition rate , short test cycle, good stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Embodiment 1, the method for preparing label
[0041] For the Ion torrent sequencing platform, the inventor provides a method for preparing tags, including the following steps:
[0042] (1) Generate all nucleotide sequences of 8bp and 10bp to form a pool of candidate tags.
[0043] (2) Screening the nucleotide sequences in the candidate tag pool, the screening criteria are as follows:
[0044] Remove the nucleotide sequence that is not C at the end of the 3' end;
[0045] Remove the nucleotide sequence whose 5' head is C;
[0046] Remove nucleotide sequences comprising AGC, GCC, AAG, CTGC, CCG, CAA, AAA, TTT, CCC, GGG;
[0047] Remove the nucleotide sequence starting from AGG at the 5' end;
[0048] Nucleotide sequences that are CC-terminated at the 3' end are removed.
[0049] (3) Perform pairwise alignment of the nucleotide sequences in the screened candidate tag pool, and remove any sequence with a difference of less than 3; use the Needleman-Wunsch algorithm fo...
Embodiment 2
[0052] Embodiment 2, label screening
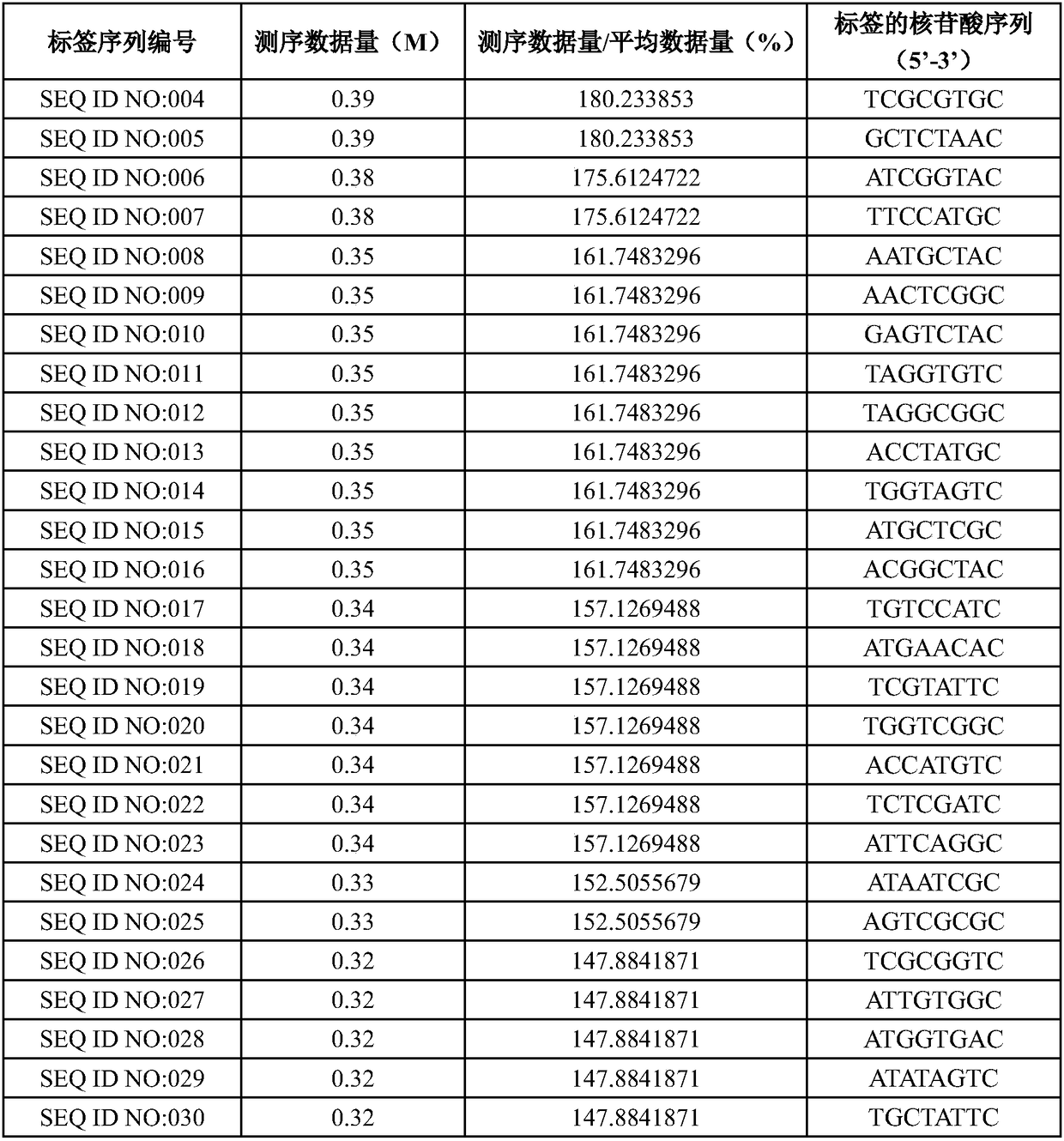
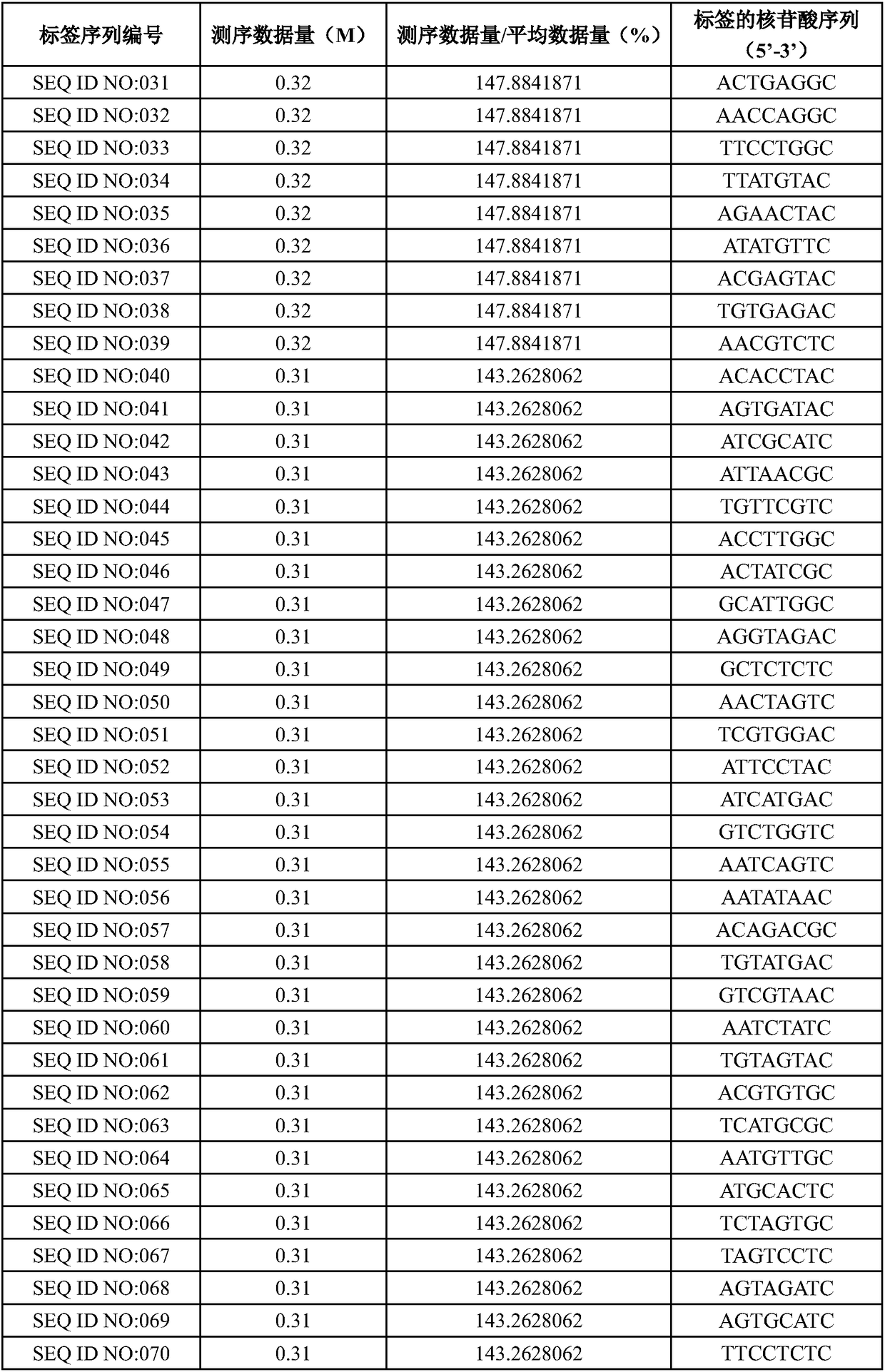
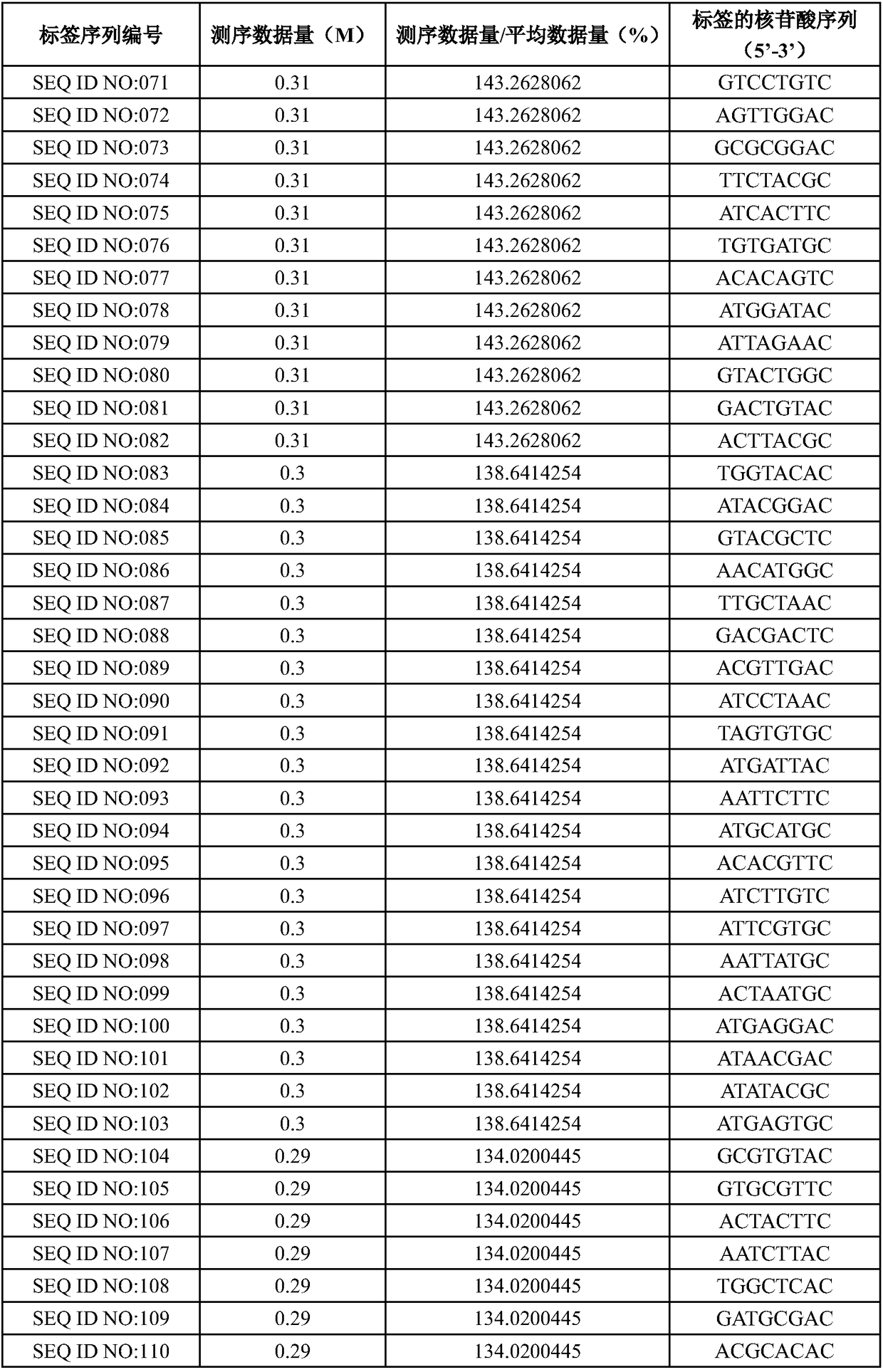
[0053] Optionally select 415 tags in the candidate tag pool obtained in step (4) of Example 1, perform test screening, use the human genome as a template, and combine the deafness gene sequencing method (refer to Example 3 for specific methods) to carry out library construction, and the 415 library High-throughput sequencing was carried out on a PI chip, and the amplification efficiency of the tag sequence was measured according to the amount of data obtained by sequencing (number of reads). The ratio of the amount of sequencing data to the average amount of data was greater than 25% as the standard, and a total of 384 samples were screened. tag sequences, and arrange the sequencing data volume in descending order, as shown in Table 1.
[0054] Table 1. Tag screening
[0055]
[0056]
[0057]
[0058]
[0059]
[0060]
[0061]
[0062]
[0063]
[0064]
[0065]
Embodiment 3
[0066] Example 3. Deafness gene sequencing method based on DNA tag library
[0067] 1. Tab connector
[0068] Using the 384 tags (hereinafter referred to as index) shown in SEQ ID NO: 004 to SEQ ID NO: 387 shown in Table 1, insert the tag upstream of the 3' end "GAT" of the tag adapter, and the tag adapter 3' end "GAT" "The universal sequence is inserted downstream, wherein the original tag linker sequence is 5'-CCATCTCATCCCTGCGTGTCTCCGACTCAGGAT-3' (SEQ ID NO: 001), and the universal sequence is: 5'-AAATGGGCGGTAGGCTTG-3' (SEQ ID NO: 002); thus constituted The new 384-index linker has the following structural form: 5'-CCATCTCATCCCTGCGTGTCTCCGACTCAG-index-GAT-AAATGGGCGGTAGGCTTG-3'.
[0069] 2. DNA extraction
[0070] A total of 1152 dried blood spot samples (400 positive samples and 752 negative samples) were selected, and the samples were scientific research samples provided by cooperative units. They were extracted using HiPure Blood DNA Mini Kit (Magen Company), and the pur...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com