DNA label sequence, sequencing library construction method and kit
A technology for labeling sequences and sequencing libraries, applied in the field of molecular biology, can solve the problems of DNA library sample loss, cumbersome operation process, and many steps of library construction, and achieve the goals of avoiding uncertain factors, simple steps, and optimizing sensitivity and repeatability Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Embodiment 1 A kind of DNA tag
[0045] In this example, on the basis of satisfying the above DNA tag design conditions, three sets of DNA tags with high specificity are designed, as shown in Table 1, Table 2, and Table 3, wherein the DNA tag sequence in Table 1 is 6 bases , the DNA tag sequence in Table 2 is 7 bases, and the DNA tag sequence in Table 3 is 8 bases, wherein, the PCR amplification primers of the same sequencing reaction system are selected from the DNA tags in the same list, and are combined with PCR The amplification primers are combined to form corresponding tag amplification primers.
[0046] Table 1 DNA tag sequence (6bp)
[0047] SEQ ID NO.
Tag sequence (5'→3')
SEQ ID NO.
Tag sequence (5'→3')
1
TAGCCA
11
GATACG
2
ACATGC
12
GTACAG
3
TGTGCA
13
GACTGT
4
ACGTTG
14
AGTGCT
5
GAGCTA
15
TACCAG
6
GTCTGA
16
TTCGCA
7
...
Embodiment 2
[0052] Example 2 Indexing PCR primers, kits for sequencing libraries constructed by PCR primer pairs and DNA tags
[0053] 1. PCR amplification primers
[0054] This example takes the detection of EGFR gene and BRAF gene mutations as an example, using the DNA tag sequence described in Example 1 and the amplification primers for the EGFR gene and BRAF gene mutation sites detected by the target to construct a sequencing library kit. That is to construct specific label PCR primers, according to the method of the present invention to determine whether there are mutations in the target detection genes of various DNA samples.
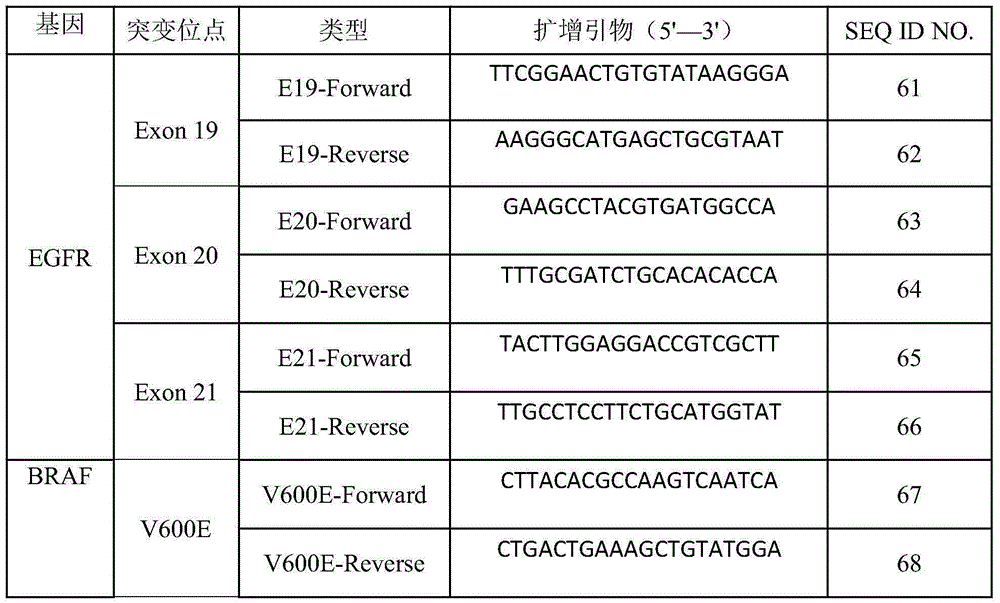
[0055] The present invention designs PCR primers for EGFR gene and BRAF gene, utilizes this set of PCR primers to carry out PCR amplification on the DNA sample, and can amplify target detection gene fragments by one-step PCR, and the specific PCR primers are:
[0056] Table 4PCR amplification primers
[0057]
[0058] 2. Labeling PCR primers and DNA labe...
Embodiment 3
[0066] Example 3 uses the labeled PCR primers in Example 2 to detect samples
[0067] 1. Sample DNA extraction:
[0068] Refer to the instructions of the AxyPrep Whole Blood Genome Mini Extraction Kit to obtain the DNA to be detected.
[0069] 2. PCR amplification of samples to be tested
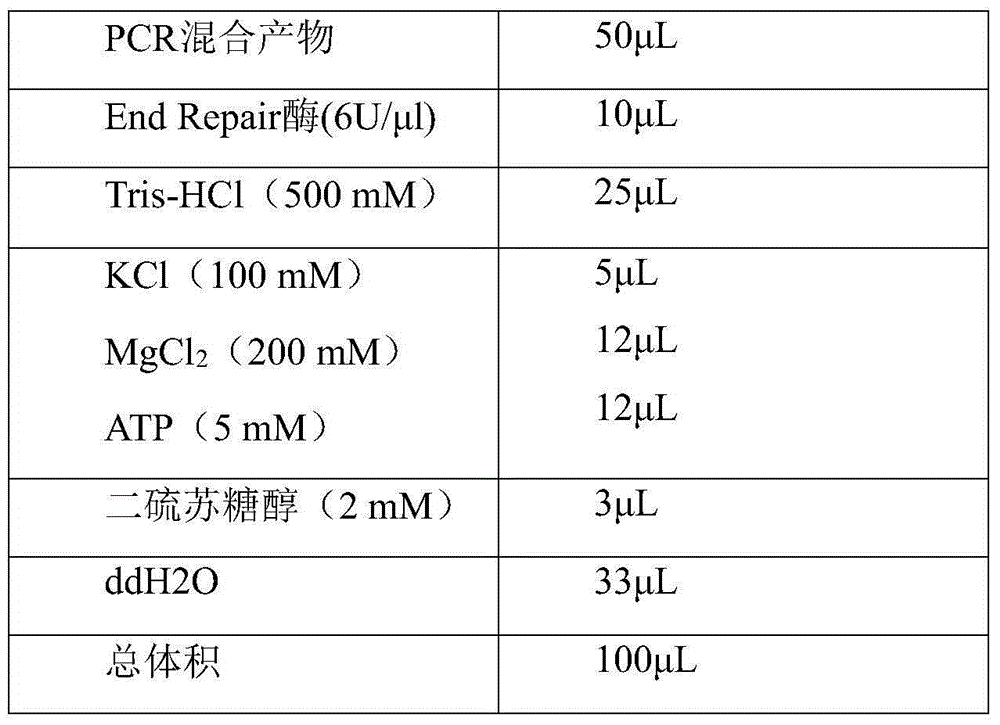
[0070] Prepare labeling primer working solution: according to the order of 20 samples in Table 5, take 100ul of PCR primer stock solution with DNA labeling in 1.5ml microcentrifuge tubes, assemble into 20 tubes of labeling PCR amplification primers, and mix well. Multiplex PCR primer working solution, for each sample, each tube of working solution contains amplification primers for EGFR gene and BRAF gene with specific DNA tags introduced at the 5' end. Amplify the target sequence containing the mutation site respectively, and the PCR reaction system is as follows:
[0071]
[0072] The PCR amplification program is: 95°C for 3min; 94°C for 20s, 56°C for 30s, 72°C for 30s, 30 cycles; 72...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com