Nav1.7 knockout mice and uses thereof
A technology of nav1.7 and mice, which is applied in the direction of recombinant DNA technology, preparations for in vivo experiments, and the use of vectors to introduce foreign genetic materials, etc., can solve problems such as feeding failure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0101] Included within the scope of the present invention are global Nav1.7 KO mice in which they have been "knocked out" or "knocked out" by insertion of a gene from the mouse (which may have a modified nucleotide sequence) or a transgene into" 1, 2 or more additional genes of interest. Such mammals can be created by repeating the methods set forth herein for generating each "knockout" or transgenic "knockin" construct or by breeding mammals each with a single gene knockout with each other, followed by screening for the presence of two knockouts. Mammals of one or more knockout and / or knockin genotypes are generated. The gene to be knocked out or knocked in can be any gene as long as at least some sequence information about the DNA to be disrupted or recombinantly expressed is available for use in making constructs and screening probes.
[0102] Knockout gene selection
[0103] Typically, the DNA to be used in a knockout construct will be one or more exon and / or intron re...
Embodiment 1
[0234] Example 1 .Backcross and outcross
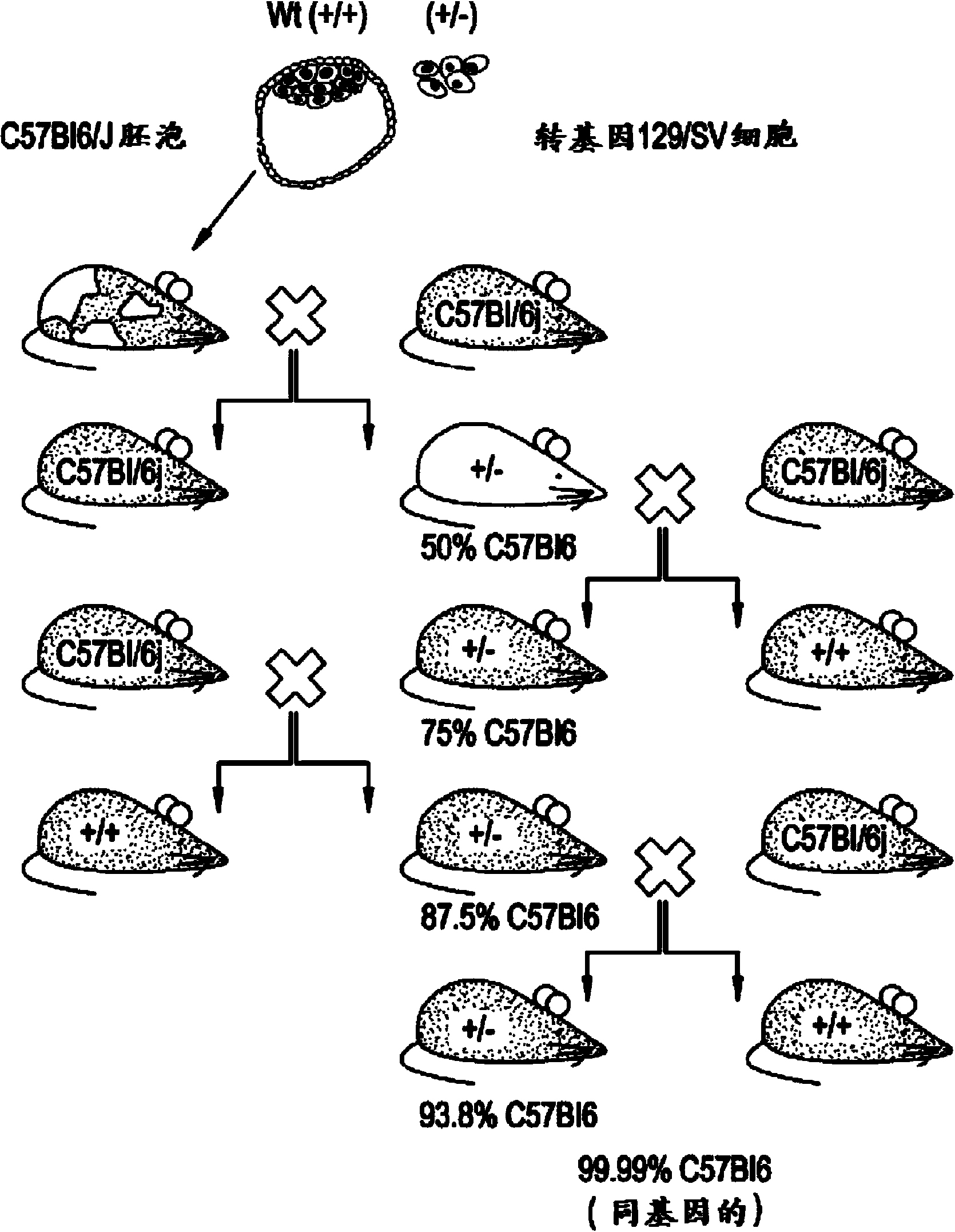
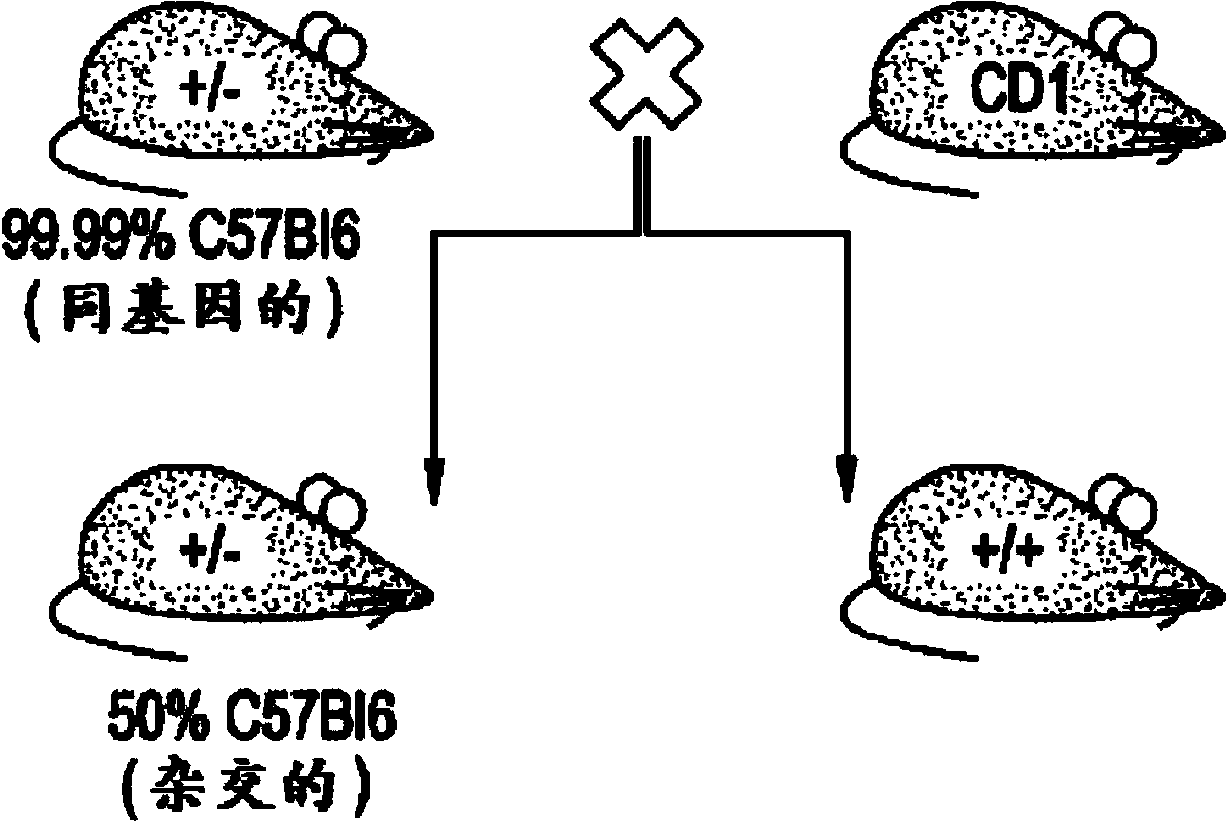
[0235] Due to the reported Na V 1.7 KO animals (heterozygous Nav1.7 from Deltagen backcrossed with C57BL / 6 for at least 8 generations, San Mateo, CA: B6.129P2-Scn9atm1Dgen / J + / - mouse) (Nassar et al., 2004), we outcrossed these animals to a CD1 background to increase the vigor of the strain, and backcrossed them to the BALB / c strain respectively to generate isogenic strains ( The first breeding pair obtained in May 2011). (see, Figure 1A -B).
[0236] No significant differences were observed in Scn9a-CD1KO or Scn9a-Balbc KO animals. We call these animals Na V 1.7 KO, and specify the strain if required. All data shown in this article were collected from a single outcross and backcross. Further backcrosses (for the BALB / c line only) are currently underway and the results obtained so far are consistent across the N4 backcross generation. No further increase in survival was observed. The final outcome of the backcross is still t...
Embodiment 2
[0259] Example 2 : Preparation of artificial mouse milk (AMA)
[0260] The method and composition of artificial mouse milk prepared was modeled closely following the protocols described by Yajima and coworkers and by Auestad and coworkers. (Yajima, M, et al., A Chemically Derived Milk Substitute that is Compatible with Mouse Milk for Artificial Rearing of Mouse Pups'Exp.Amin.55(4), 391-397, (2006); Auestad, N, et al., Milk-substitutes comparable to rat's milk; their preparation, composition and impact on development and metabolism in the artificially reared rat, Br.J.Nutr.61:495-518(1989)).
[0261] Reagents and methods. Reagents used, including sources, amounts used, solvents and amounts used, and various notes are listed in Table 3 (below).
[0262] method. Step 1: Weigh the sodium hydroxide and transfer it to a 10-L beaker. Distilled water (1.3 L) was added and an overhead stirrer was installed. Potassium hydroxide was weighed and added to this solution, and the sa...
PUM
| Property | Measurement | Unit |
|---|---|---|
| impedance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com