Method and kit for simultaneous detection of human coronavirus 229E, OC43 and NL63
A technology of human coronavirus and coronavirus, applied in the field of biotechnology applications, can solve the problems of difficult detection of trace nucleic acid and accurate quantification, long time-consuming, cumbersome operation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1. Human coronavirus 229E, OC43, NL63 corresponding N gene-specific primers and probe design and synthesis
[0032] 1.1 Selection of the corresponding target gene N gene and determination of the conserved region of representative strains of human coronavirus 229E, OC43, and NL63
[0033] The sequences used in this study were selected from NCBI GenBank (bttp: / / www.ncbi.nlm.nih.gov), the main selection basis is: (a) strains with a relatively recent age; (b) full-length sequences and main sequences (c) Select a representative strain after comparing the sequence with the subtype. The information of the corresponding N genes of the selected human coronavirus 229E, OC43, and NL63 representative strains is shown in Tables 1-1, 1-2, and 1-3.
[0034] In this study, the corresponding N genes of the selected representative strains of human coronavirus 229E, OC43, and NL63 were input into the DNAssist software for homologous comparison, and the sequence conserved regio...
Embodiment 2
[0047] Example 2. The establishment of a one-step multiple fluorescent RT-PCR detection system for human coronavirus 229E, OC43, and NL63
[0048] 2.1 According to Invitrogen's nucleic acid extraction kit ( Viral RNA Mini Kit) to extract sample nucleic acid.
[0049] 2.2 Preparation of human coronavirus 229E, OC43, NL63 one-step multiplex fluorescent RT-PCR detection system
[0050] Ambion’s AgPath-IDTM One-step RT-PCR Kit (P / N: 4387424) was used to prepare reagents, and the system was 50 μl:
[0051] 2×RT-PCR buffer 25μl
[0052] 25×RT-PCR Enzyme 2μl
[0053] Detection Enhancer 3μl
[0054] Primers and probes are added at the same time as shown in SEQ NO: 1, 2, 4, 5, 7, 8, 10, 11
[0055] 0.5 μl of each primer, the final primer concentration is 200nM; add such as SEQ NO: 3,
[0056] 0.5 μl each of the probe primers indicated in 6, 9, and 12, and the final concentration of the probe primers is 100 nM
[0057] Template 6 μ...
Embodiment 3
[0068] Example 3. Specific identification of human coronavirus 229E, OC43, NL63 one-step multiple fluorescent RT-PCR detection method
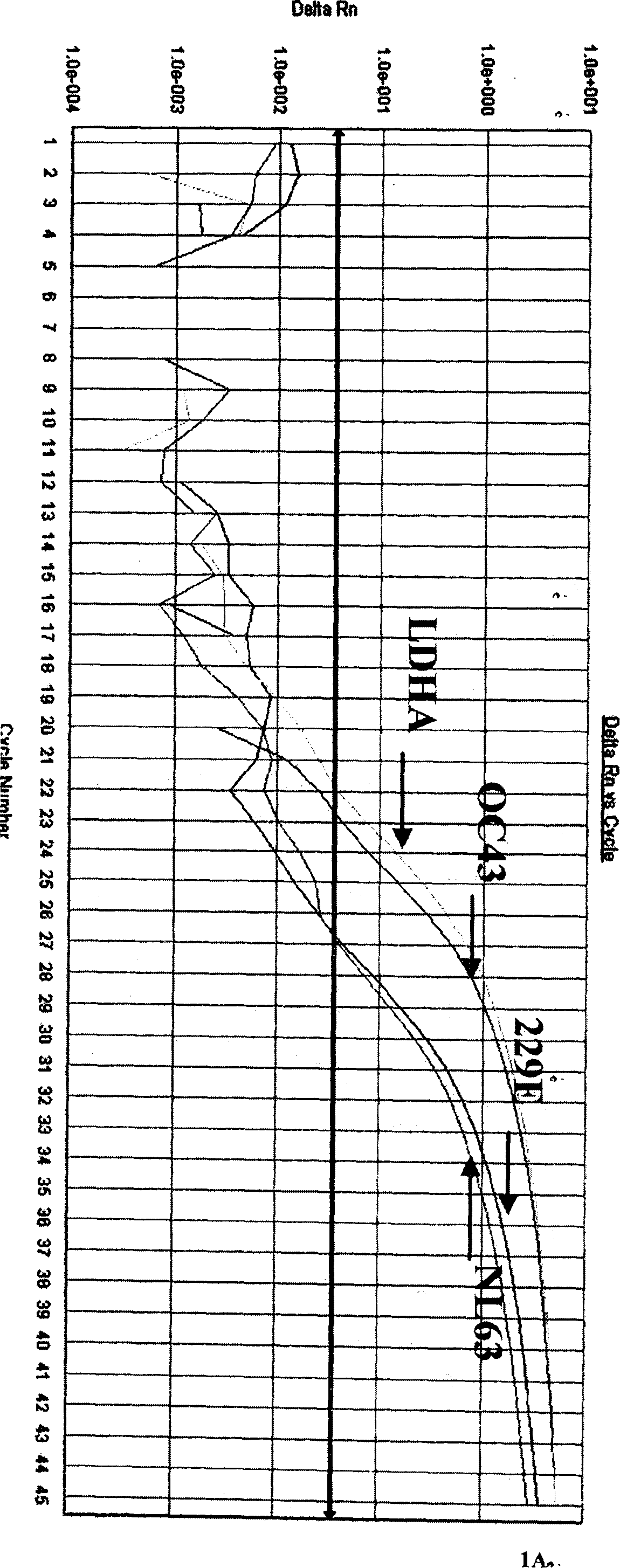
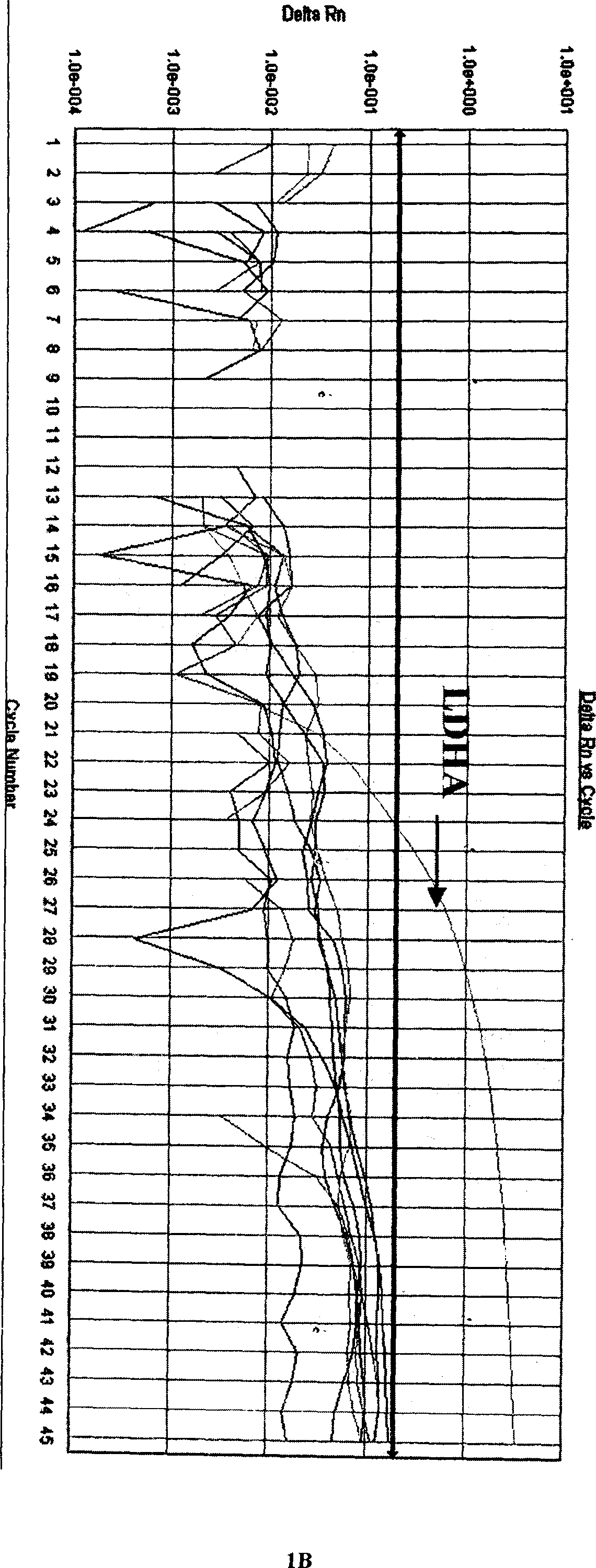
[0069] Utilize the multiplex fluorescent RT-PCR reaction system that embodiment 2 establishes respectively to human coronavirus 229E, human coronavirus OC43, human coronavirus NL63, influenza A virus, influenza B virus, influenza C virus, parainfluenza The positive nucleic acids of virus, respiratory syncytial virus, enterovirus, adenovirus, and boca virus were detected, and the results showed that only human coronavirus 229E, OC43, and NL63 showed corresponding specificity in the detection channels of FAM, JOE, and ROX, respectively. There was no cross-reaction, but the other 8 viruses had no amplification curve except the internal quality control CY5 channel, and the other three channels had no amplification curve, indicating that the method has strong specificity (see Fig. 1).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com