Real-time fluorescence multi-RT-PCR (Reverse Transcription-Polymerase Chain Reaction) method for simultaneously detecting human coronaviruses 229E, OC43, NL63 and HKU1
A RT-PCR, coronavirus technology, applied in microorganism-based methods, biochemical equipment and methods, DNA/RNA fragments, etc., can solve problems such as high sensitivity real-time quantification, which is not conducive to early diagnosis of diseases
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0143] Primer design and screening
[0144] Download the existing sequences of human coronavirus 229E, OC43, NL63 and HKU1 in Genebank, select the M gene of 229E, the N gene of OC43, the N gene of NL63 and the N gene of HKU1 as the target genes for detection, using MEGA software Multiple sequence alignments were carried out to find out the respective conserved regions.
[0145]According to the comparison results, 12 pairs of primers were designed in the conserved region of each target gene using "NCBI / Primer-BLAST", including 3 pairs of primers for amplifying the M gene of 229E (SEQ ID NO.: 1, 2, 19-22) , 4 pairs of primers (SEQ ID NO.: 9, 10, 23-28) for amplifying the N gene of OC43, 1 pair of primers for amplifying the N gene of NL63 (SEQ ID NO.: 5, 6) and 4 pairs of primers for amplifying the N gene of NL63 Primers for increasing the N gene of HKU1 (SEQ ID NO.: 7, 8, 29-34). All primers are analyzed by "Primer-BLAST" to ensure that there is no cross-homology with other nu...
Embodiment 2
[0156] Modification of OC43 primer
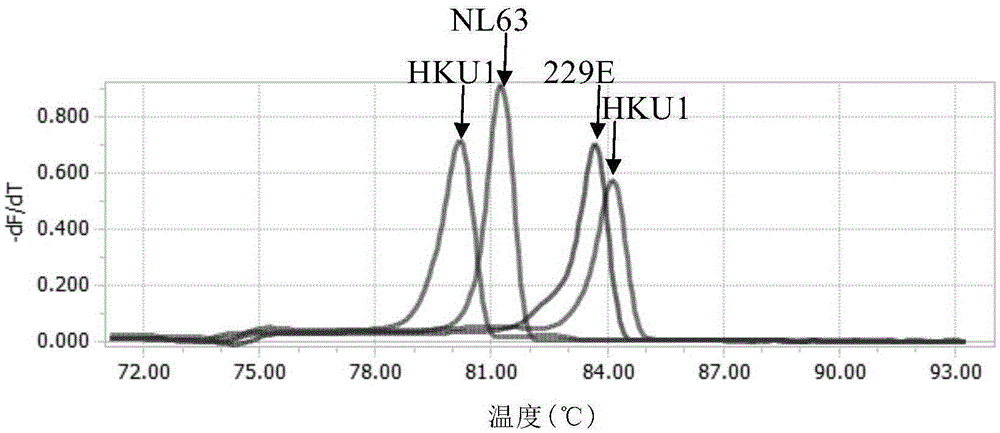
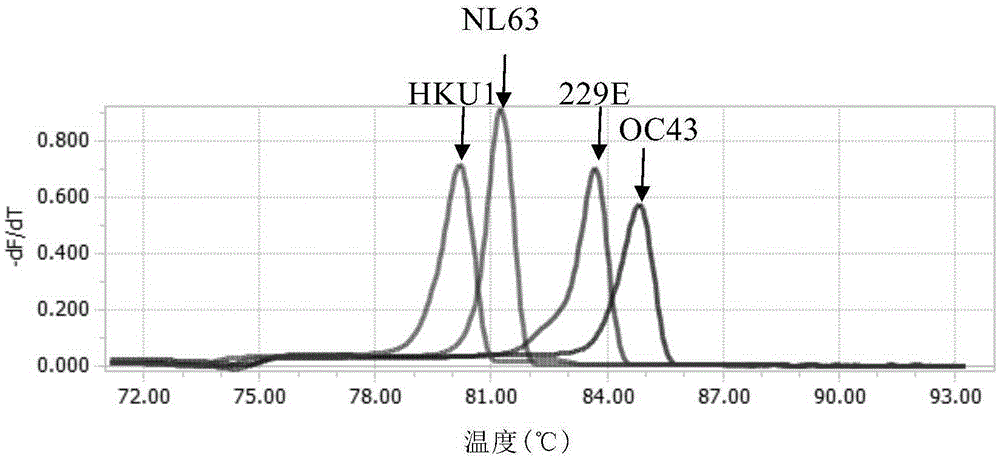
[0157] From the results of melting curve analysis of the four templates amplified by the best primer combination in Example 1, it can be seen that the difference in Tm between 229E and OC43 is less than 1°C. In the absence of a better primer scheme , try to modify the primers of OC43 as follows:
[0158] Independently select a GC-rich sequence containing 9 bases: ATCCCGGGG, use the "Scrambled DNA Sequence Tool" in the "Sequence Processing Online Toolkit (SMS)" to generate a series of sequences with the same base composition as the fragment For fragments with different base sequencing, try to add the 5' ends of the OC43 amplification primers (SEQ ID NO: 9, 10) selected in Example 1, respectively.
[0159] All modified primer sequences were analyzed using the software "Vector NTI", and the modified primers that formed primer-dimers and less hairpin structures were screened out, and the optimized modified sequences were as follows:
[0160] ...
Embodiment 3
[0165] RNA in vitro transcription
[0166] The T7 promoter sequence: TAATACGACTCACTATAG was added to the 5' ends of the upstream primers of the four viruses (see sequence table SEQ ID NO.: 1, 5, 7 and 9), and the modified primer sequence was named T7-229E-F ( SEQ ID NO.:11), T7-OC43-F (SEQ ID NO.:12), T7-NL63-F (SEQ ID NO.:13), T7-HKU1-F (SEQ ID NO.:14).
[0167] The four viruses were amplified using the above primers in combination with their corresponding downstream primers (see SEQ ID NO.: 2, 6, 8, 10 in the sequence listing).
[0168] After the amplified product is recovered and purified, it can be used as a template for in vitro transcription.
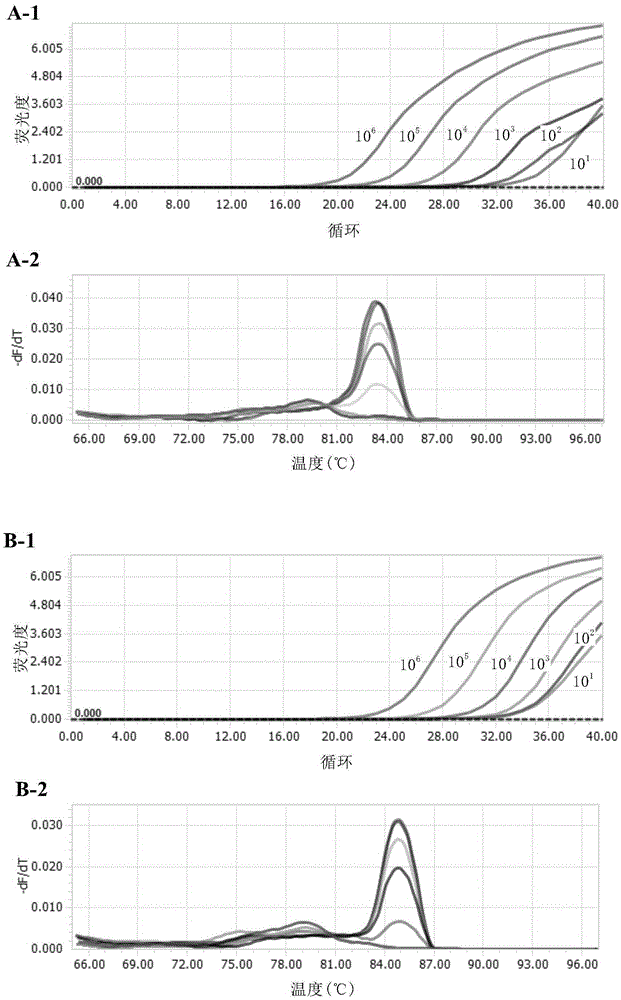
[0169] Promega's "RiboMAX Large Scale RNA ProductionSystem-T7" kit was used for in vitro transcription, and the reaction process used the method provided by the reagent manufacturer. The concentration and purity of RNA products transcribed in vitro were measured using Nanodrop, and the unit was converted into copies / μL. To avoi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com