Application of Pacific oyster C-type lectin-2(CgCLec-2) recombinant protein with antibacterial activity
A technology of antibacterial activity and recombinant protein, applied in the field of molecular biology, can solve problems such as economic loss
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] The in vitro prokaryotic recombinant expression of the long oyster lectin-2 (CgCLec-2) gene coding region comprises the following steps:
[0022] 1. Construction of recombinant vector
[0023] The recombinant vector used in the present invention is pET-32a prokaryotic expression vector.
[0024] By PCR technology, gene-specific primers with specific restriction sites of Bam H I and Hind III were added at the 5' ends to amplify the coding region fragment of C-type lectin CgCLec-2 of oyster oyster:
[0025] Gene-specific primers are:
[0026] P1 (5'-CGCGGATCCATGGATACAGAAGTGACCTCAT-3'):
[0027] P2(5'-CCCAAGCTTCTATAAAGTTTGTTCACAGATG-3'),
[0028] The reaction conditions were as follows: 94°C pre-denaturation for 5 minutes, followed by the following cycles: 94°C denaturation for 30 seconds, 55°C annealing for 30 seconds, 72°C extension for 45 seconds, a total of 35 cycles, and finally 72°C extension for 10 minutes. The amplified fragment was purified and recovered, and ...
Embodiment 2
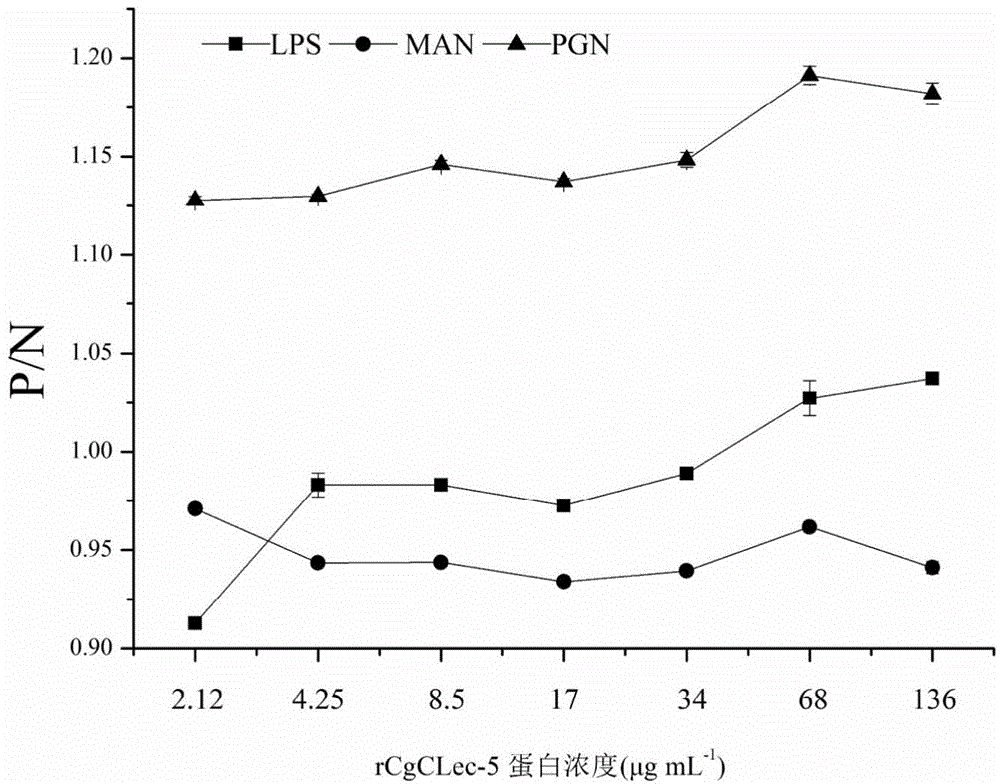
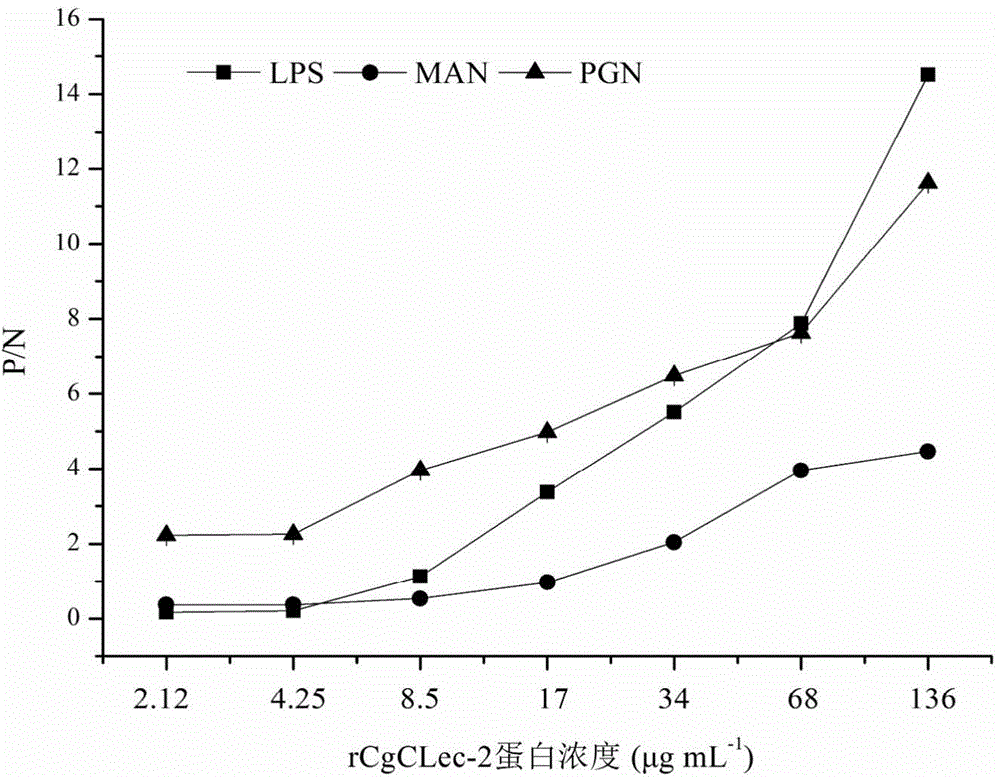
[0051] Long oyster recombinant protein rCgCLec-2 recognizes and binds carbohydrates in a calcium-dependent manner
[0052] 20 μg of LSP, PGN and MAN were dissolved in 50 mM sodium carbonate-sodium bicarbonate buffer (pH 9.6), 100 μL per well was coated with a microtiter plate, and left overnight at 4°C. In order to avoid non-specific binding, 200 μL of 3% BSA was added and blocked for 1 h at 37 °C. Afterwards, 100 μL of 2-fold serially diluted recombinant protein rCgCLec-2 was added to each well, and the final concentration was 10 mM Ca 2+ or without adding Ca 2+ Incubate at 18°C for 3 hours. Add 100 μL rat anti-rCgCLec-2 polyclonal antibody (1:1000) diluted with 3% BSA, and incubate at 37° C. for 1 h. Add 100 μL of HRP-labeled goat anti-rat IgG (1:3000), and incubate at 37°C for 1 hour. Wash 3 times with TBST between each incubation step, 5 min each time. After secondary antibody incubation and thorough washing, 100 μL TMB was added to each well, and after 30 minutes o...
Embodiment 3
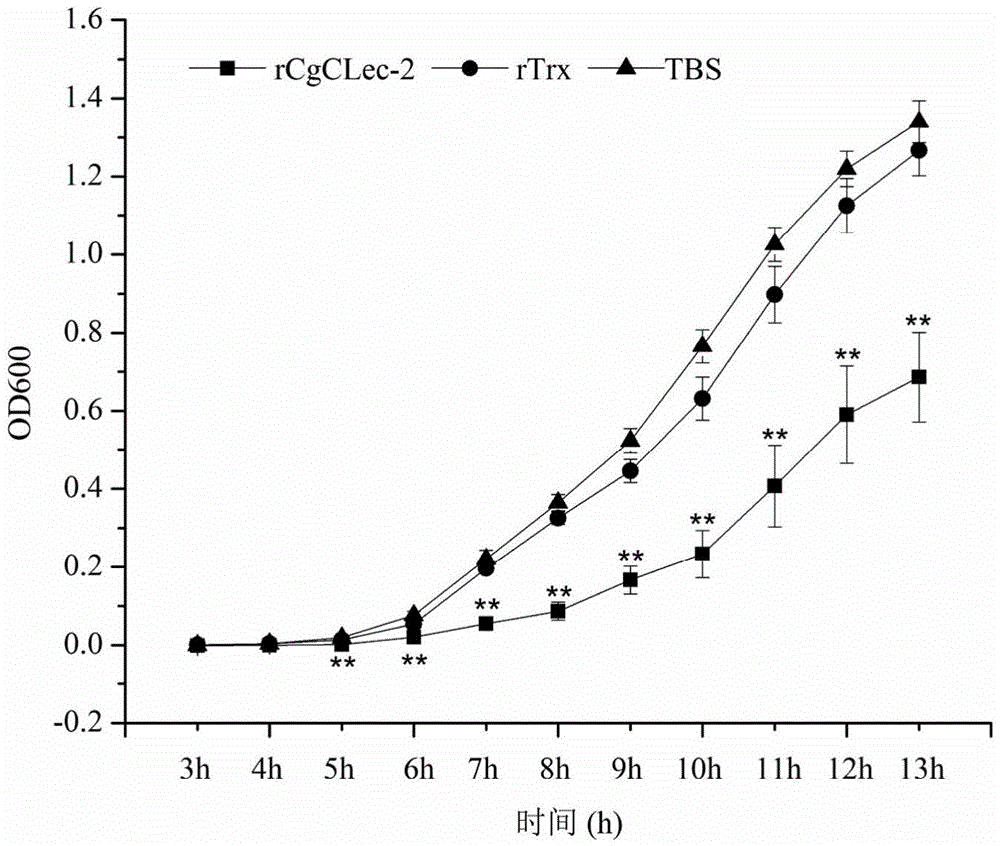
[0054] Antibacterial activity of recombinant protein rCgCLec-2 from oyster oyster
[0055] 1. Culture and preparation of microorganisms
[0056] Use LB medium for Staphylococcus aureus at 37°C, 220rpm, and when it reaches the logarithmic growth phase, dilute the bacteria with Tris-HCl (50mmol L-1, pH=8.0) so that the number of colonies per milliliter of the bacteria solution is about It is 1×103CFU.
[0057] 2. Determination of antibacterial activity of recombinant protein CgCLec-2
[0058] Staphylococcus aureus in the logarithmic growth phase was collected by centrifugation and washed and resuspended with TBS (50mM Tris-Hcl, 150mM NaCl) (10 4 CFU). 50 μL of long oyster recombinant protein CgCLec-2 with an equal volume of Staphylococcus aureus suspension in CaCl 2 Incubate at room temperature for 2 h at a final concentration of 10 mM. Take 20 μL of the above mixture in a flat-bottomed 96-well plate (Costar, Fisher), add 200 μL LB liquid medium to each well, shake and cult...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com