A method for detecting the source of miRNA
A source, technology to be tested, used in the fields of bioinformatics and genomics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
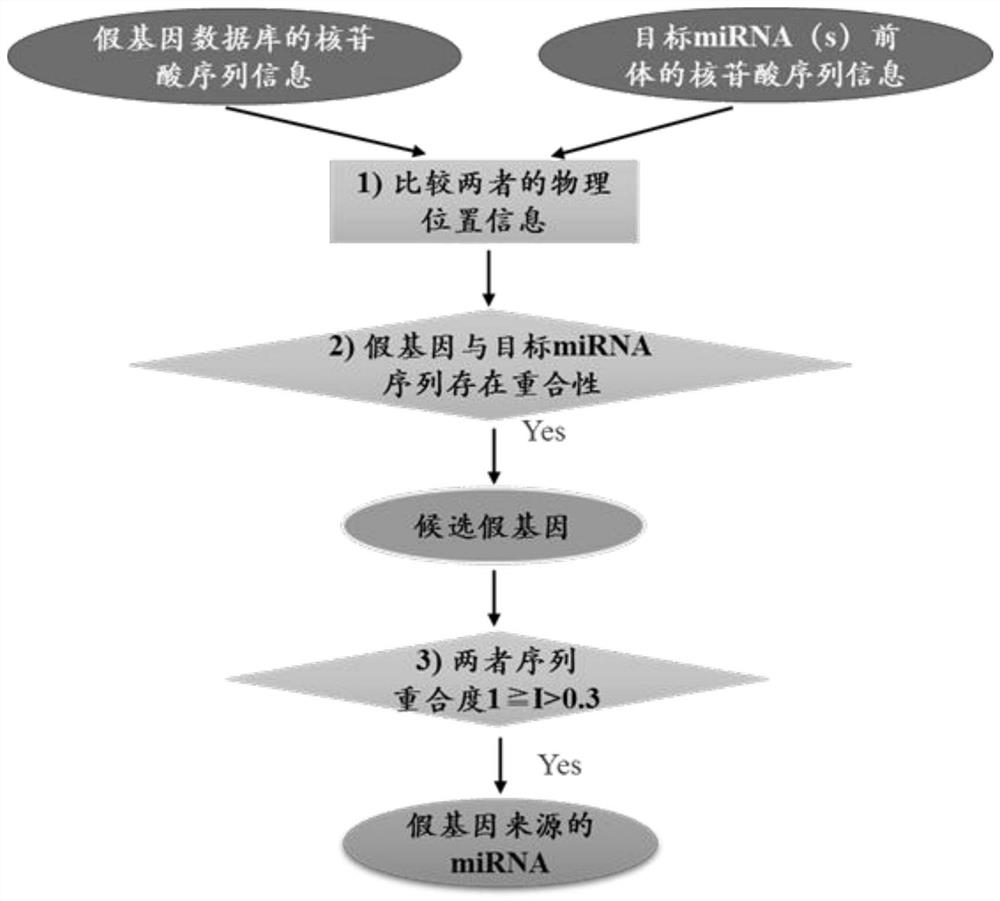
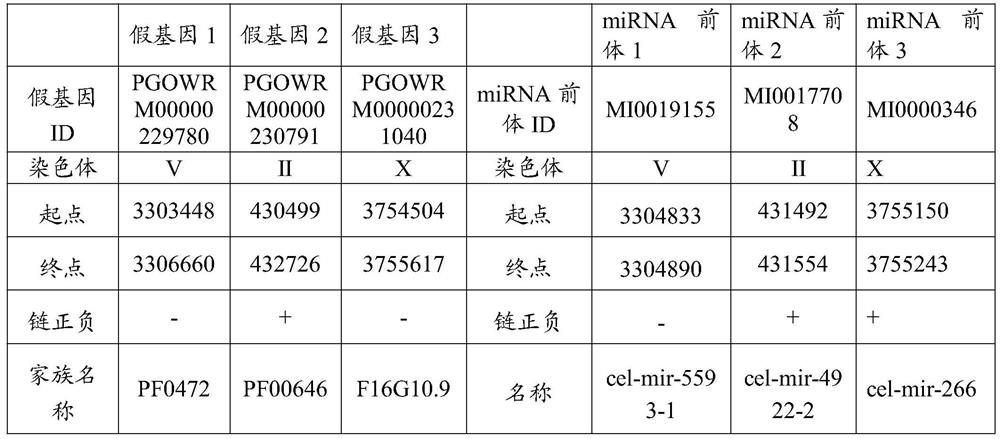
[0038] Using an approach to investigate the relationship between pseudogenes and miRNAs, a genome-wide database of known miRNA precursors and pseudogenes were correlated in the C. elegans elegans.
[0039] S1, by querying the miRNA database miRBase website (http: / / mirbase.org / index.shtml) to obtain information about all known target miRNAs precursor sequences in Caenorhabditis elegans, including their chromosome location The physical location, name and the positive or negative of the DNA strand where it is located, etc.;
[0040] S2, by querying the pseudogene database PseudoFam website (http: / / pseudofam.pseudogene.org / pages / main / about.jsf) to obtain the relevant information of the Caenorhabditis elegans genome pseudogene database, including its physical location on the chromosome, The name and the positive or negative nature of the DNA strand where it resides, etc.;
[0041] S3, compare the physical location of the target miRNA(s) of C. elegans with the pseudogene sequence o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com