Stromal gene signatures for diagnosis and use in immunotherapy
A technology of immunotherapy and matrix, applied in the direction of disease diagnosis, immunoglobulin, chemical instruments and methods, etc., which can solve the problems of not experiencing therapeutic benefits
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0338] The present invention provides following embodiment:
[0339] 1. A method for treating an individual with a disease or condition, the method comprising:
[0340] a) determining the presence of a matrix gene signature comprising one or more of FAP, FN1, MMP2, PDGFRB, or THY1 in a sample from the individual, wherein the matrix gene signature is one or more of the genes An increase in the expression level relative to the median level identifies a treated individual; and
[0341] b) administering to said individual an effective amount of immunotherapy and an inhibitory stroma antagonist.
[0342] 2. A method for improving immunotherapy in an individual with a disease or condition, the method comprising:
[0343] a) determining the presence of a matrix gene signature comprising one or more of FAP, FN1, MMP2, PDGFRB, or THY1 in a sample from the individual, wherein the matrix gene signature is one or more of the genes An increase in the expression level relative to the med...
Embodiment 1
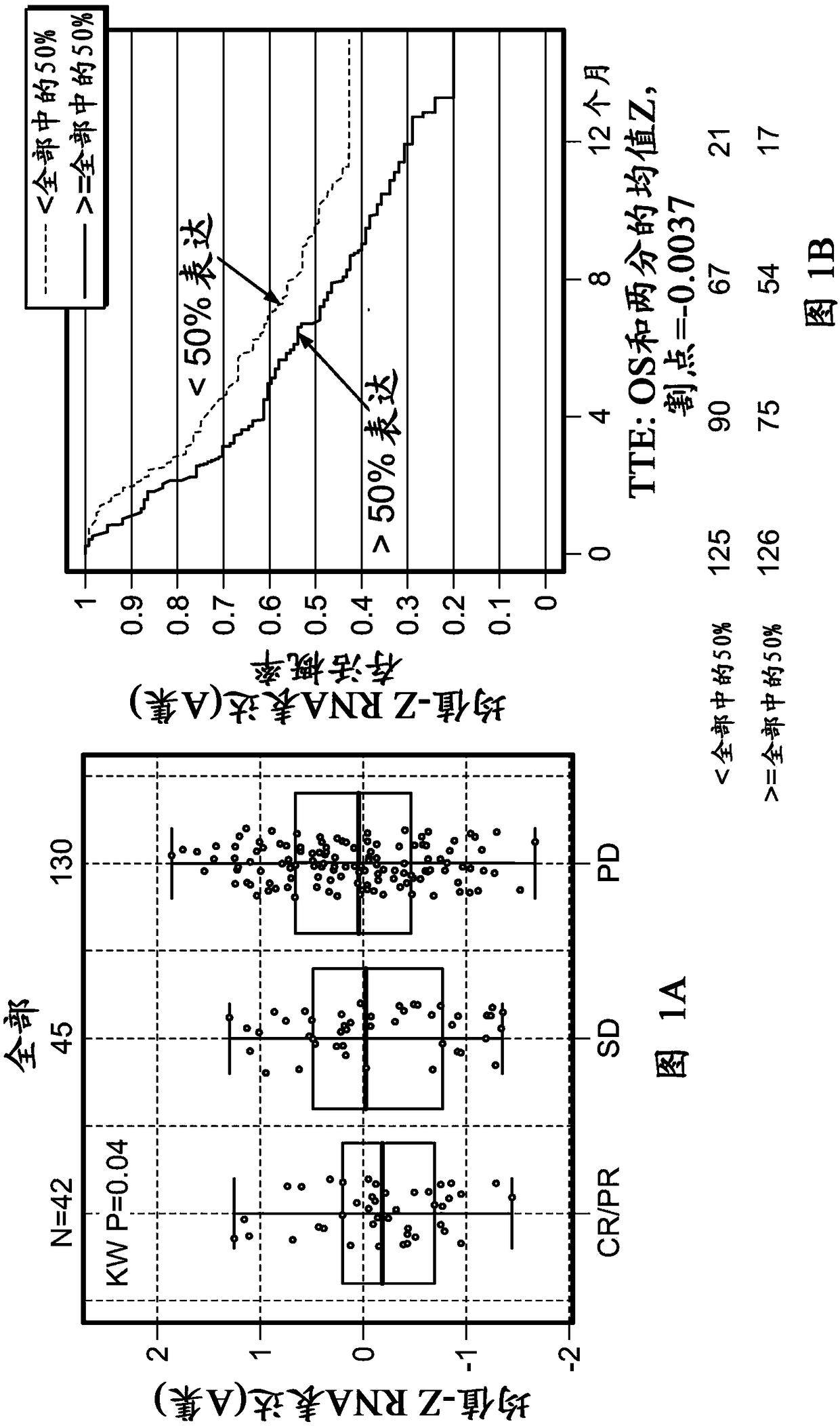
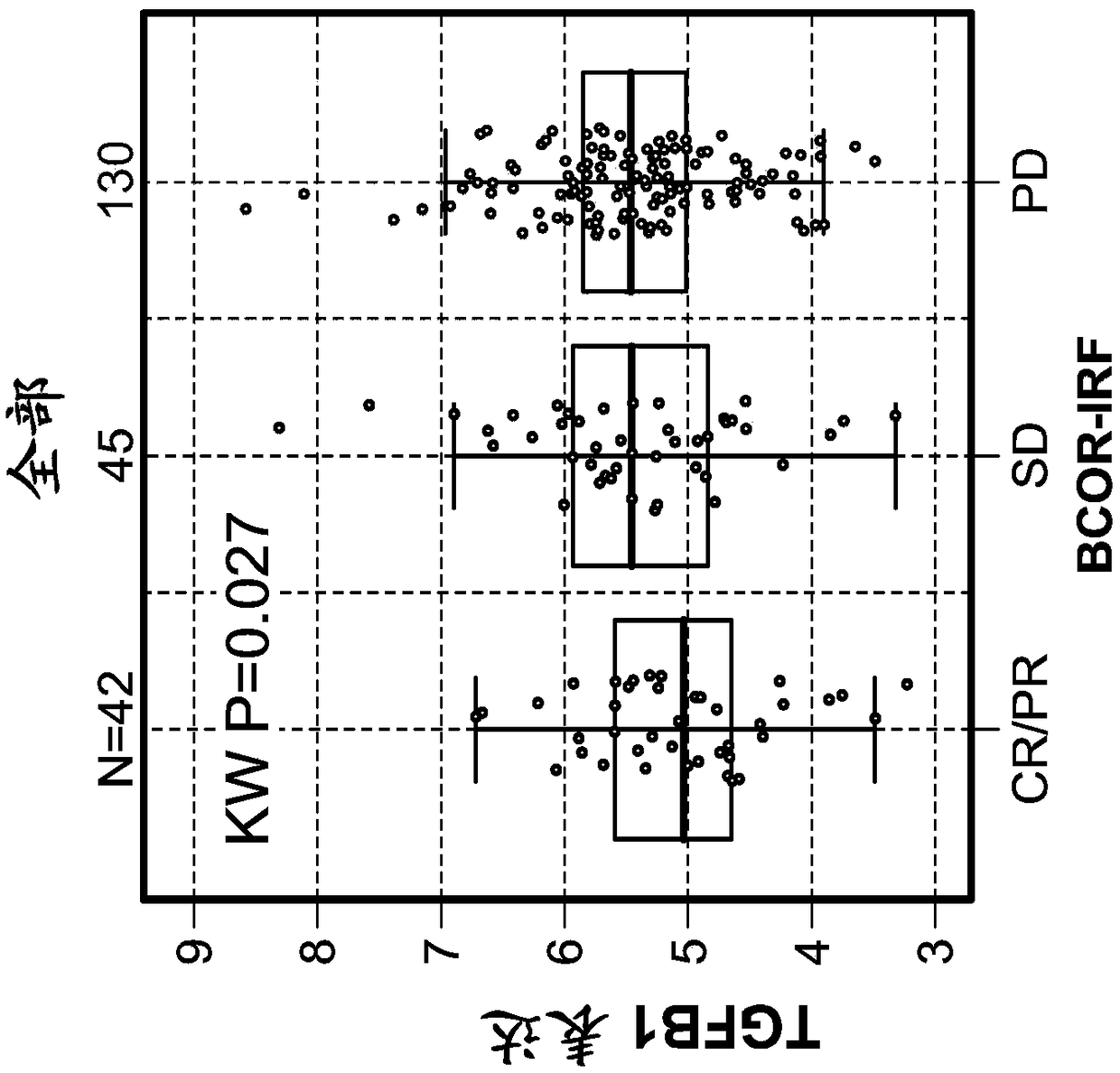
[0508] Example 1: Tumor-infiltrating stromal gene signatures across urothelial carcinoma (UC) and their association with resistance to anti-PD-L1 antibody therapy
[0509] foreword
[0510] To assess and understand the complexity of factors that may regulate or suppress antitumor immunity and thus contribute to resistance to immunomodulatory therapies, a highly sensitive immune gene expression assay was implemented to investigate Tumor microenvironment (TME) of tumor tissue before treatment.
[0511] Materials and methods
[0512] The Illumina TruSeq RNA Access RNA-seq kit was implemented to investigate the tumor microenvironment (TME) in pre-treatment tumor tissues from urothelial carcinoma patients (n=217). Illumina TruSeq RNA Access RNA-seq captures and investigates genes across the entire human genome (>20,000 genes).
[0513] RNA was extracted from formalin-fixed, paraffin-embedded archived tissue. Tumor tissue was collected clinically from the Phase II IMvigor210 stu...
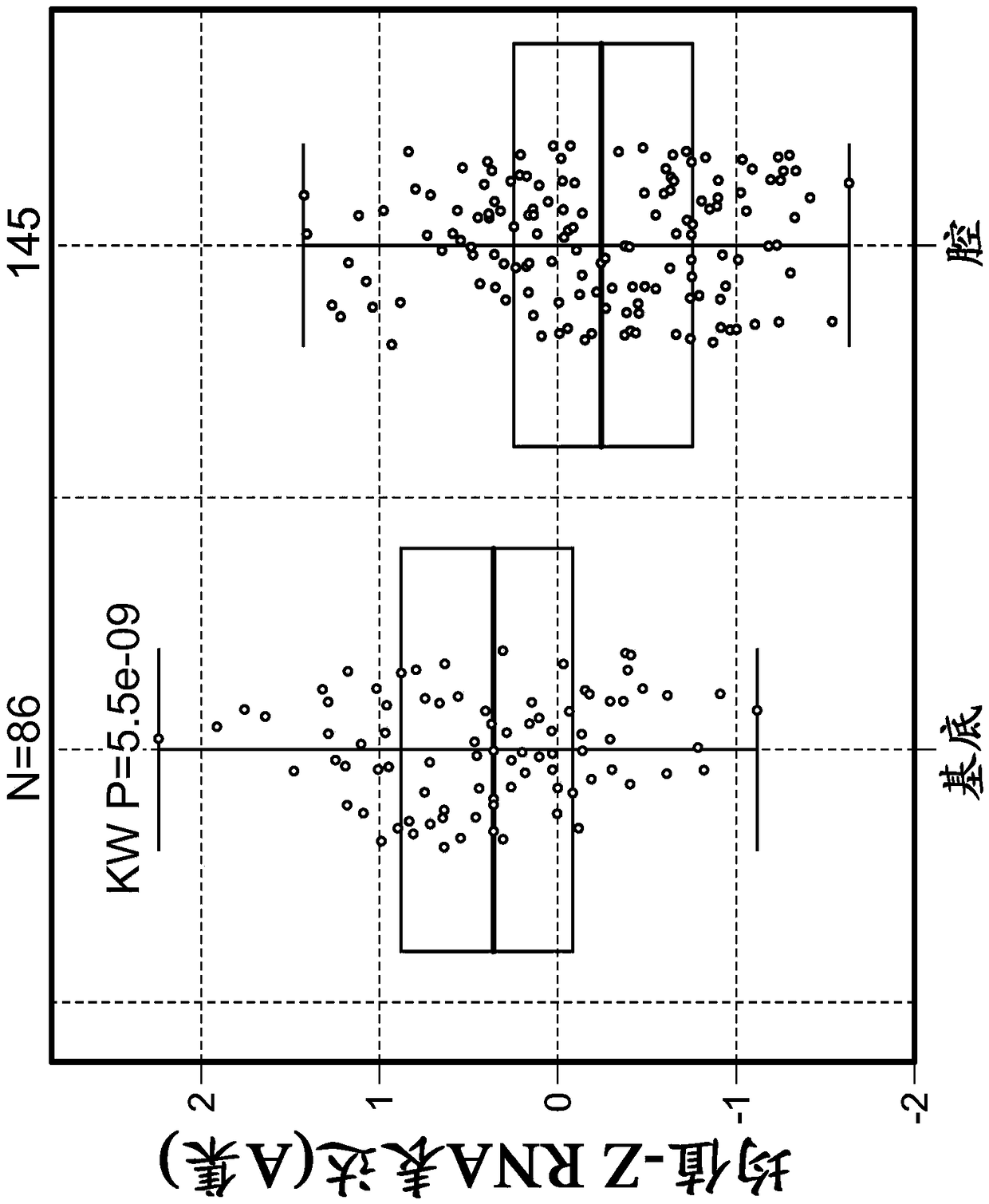
Embodiment 2
[0521] Example 2: Tumor stromal signatures across five cancer types and their association with disease prognostic factors Introduction
[0522] To further assess and understand the complexity of factors that may regulate or suppress antitumor immunity and thus contribute to response or resistance to immunomodulatory therapies, a highly sensitive immune gene expression assay was implemented to investigate (BC), tumor microenvironment (TME) of pretreatment tumor tissues of lung cancer, melanoma, RCC, and bladder cancer.
[0523] Materials and methods
[0524] Implementation of the Illumina TruSeq RNAAccess RNA-seq kit to investigate tumors from BC (n=73), lung (n=59), melanoma (n=34), RCC (n=55), and bladder cancer (n=44) The tumor microenvironment (TME) of the patient's tumor tissue before treatment. Illumina TruSeq RNAAccess RNA-seq captures and investigates genes across the entire human genome (>20,000 genes).
[0525] RNA was extracted from formalin-fixed, paraffin-embedd...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com