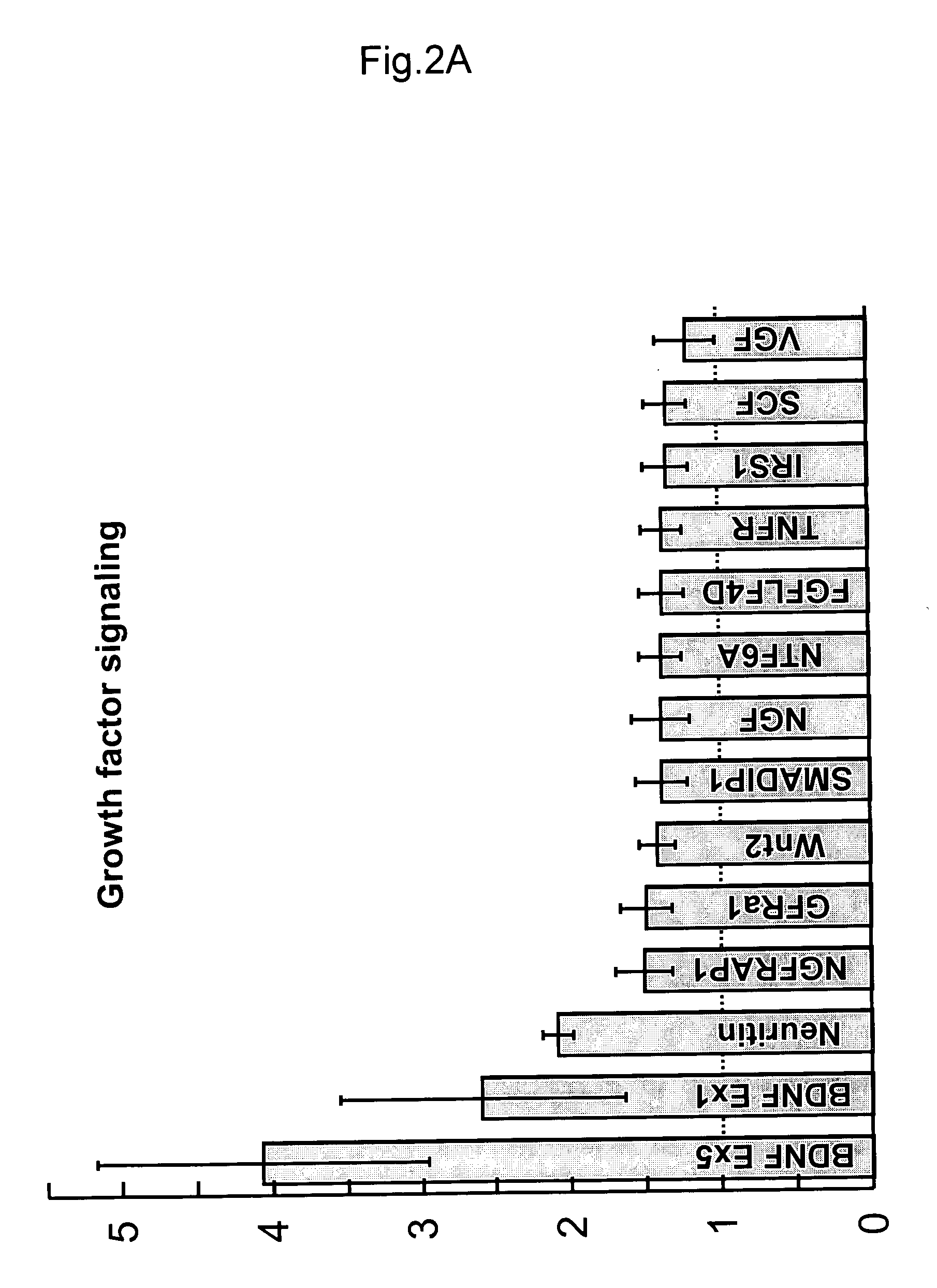
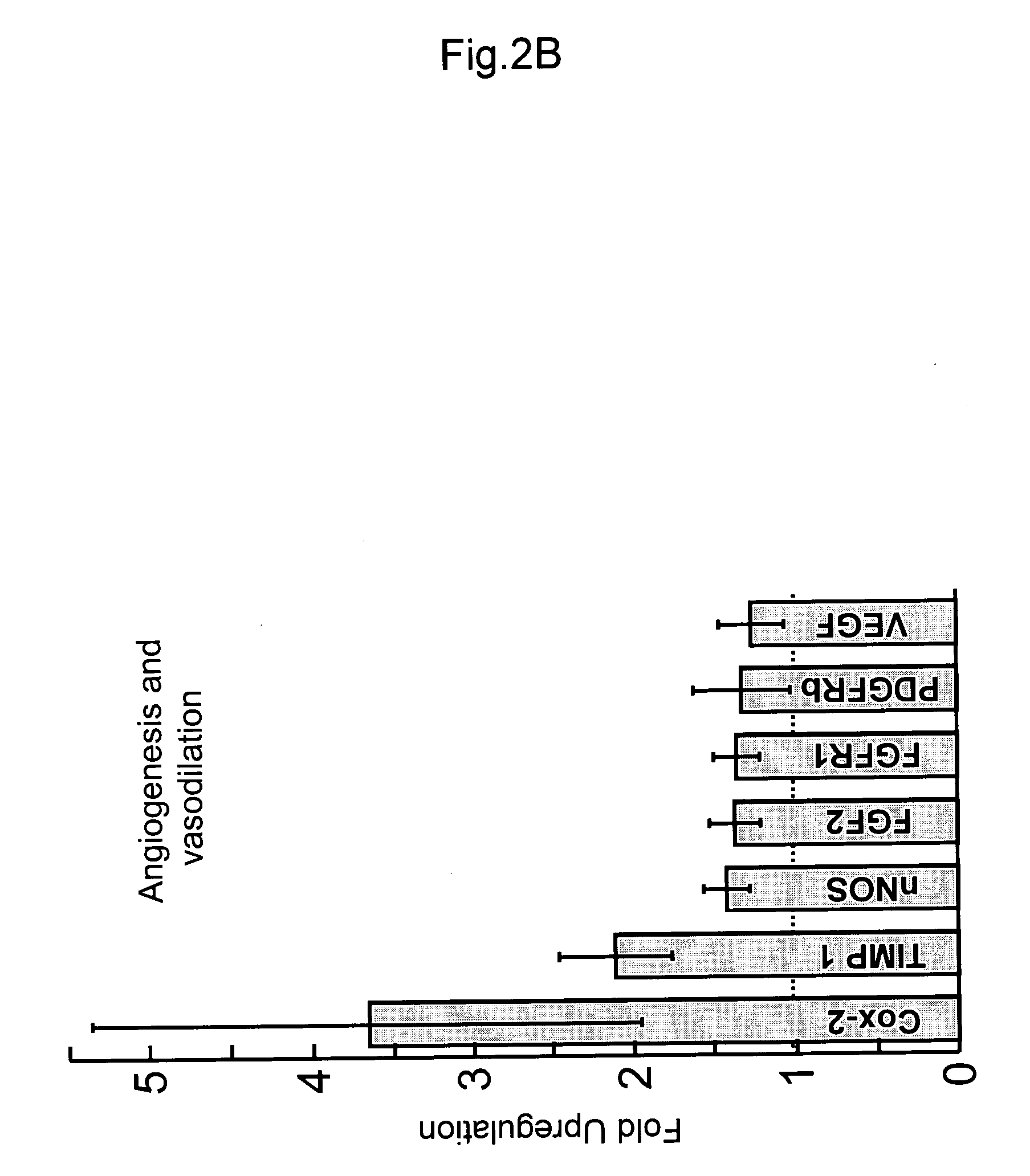
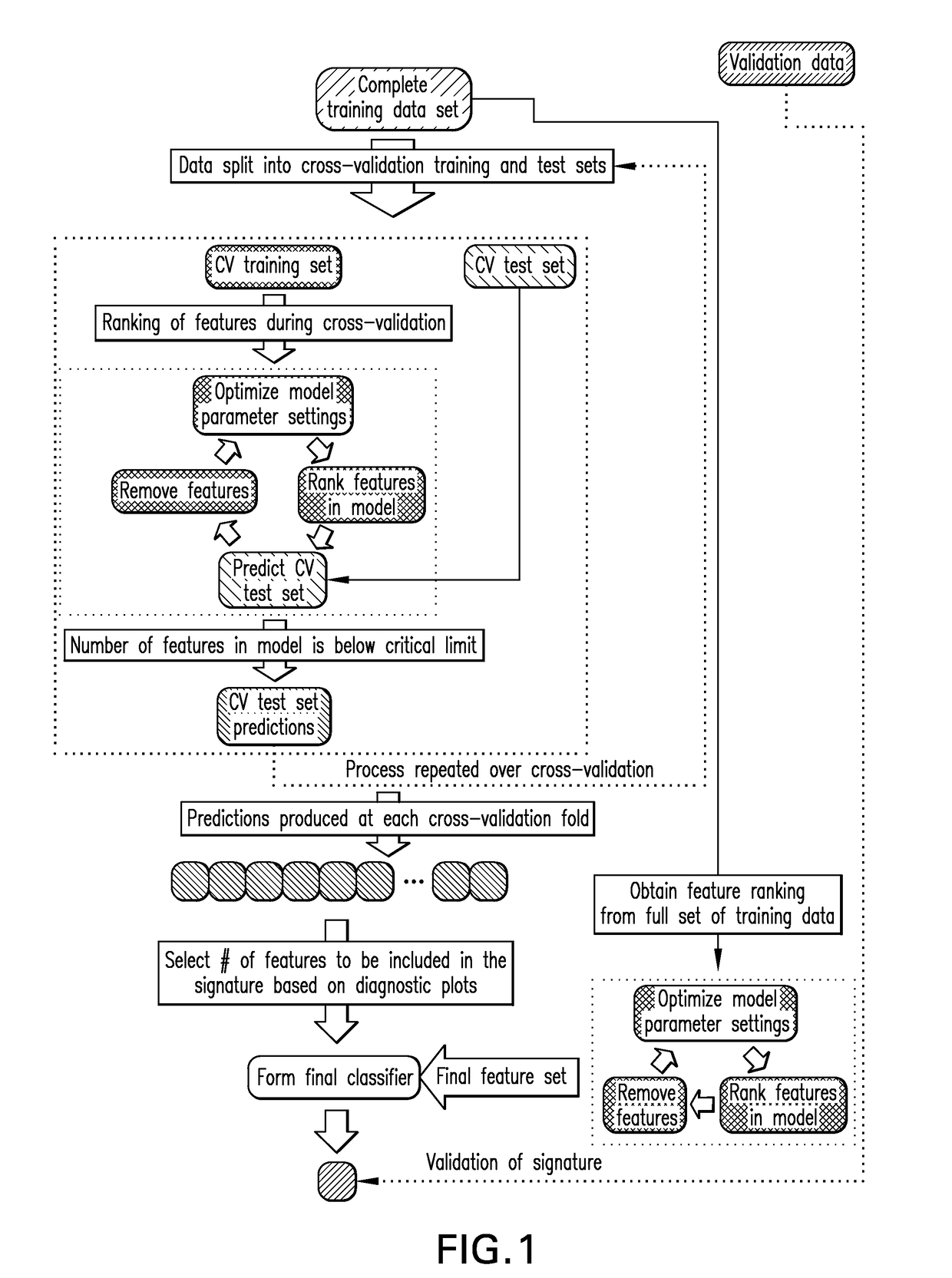
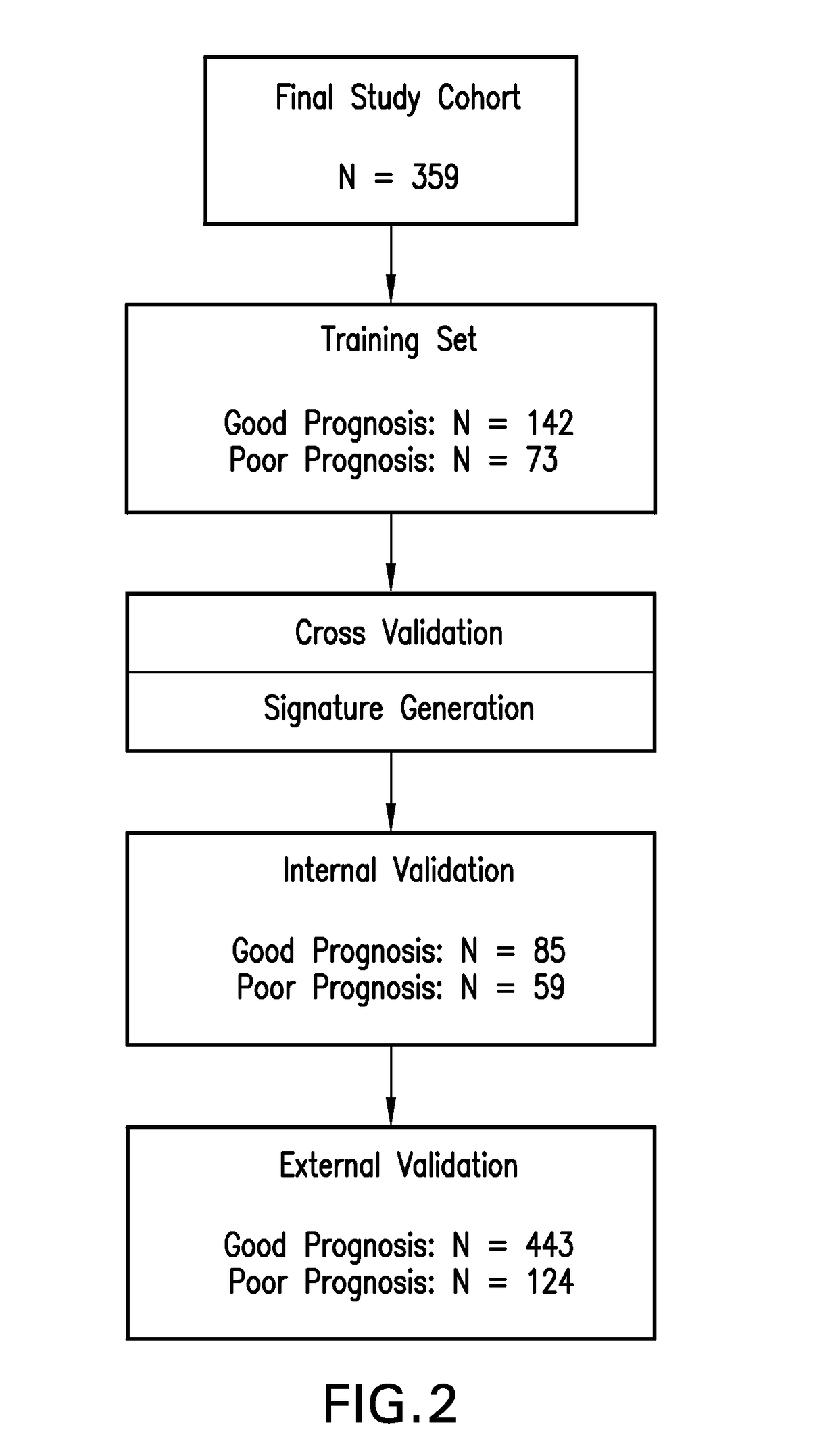
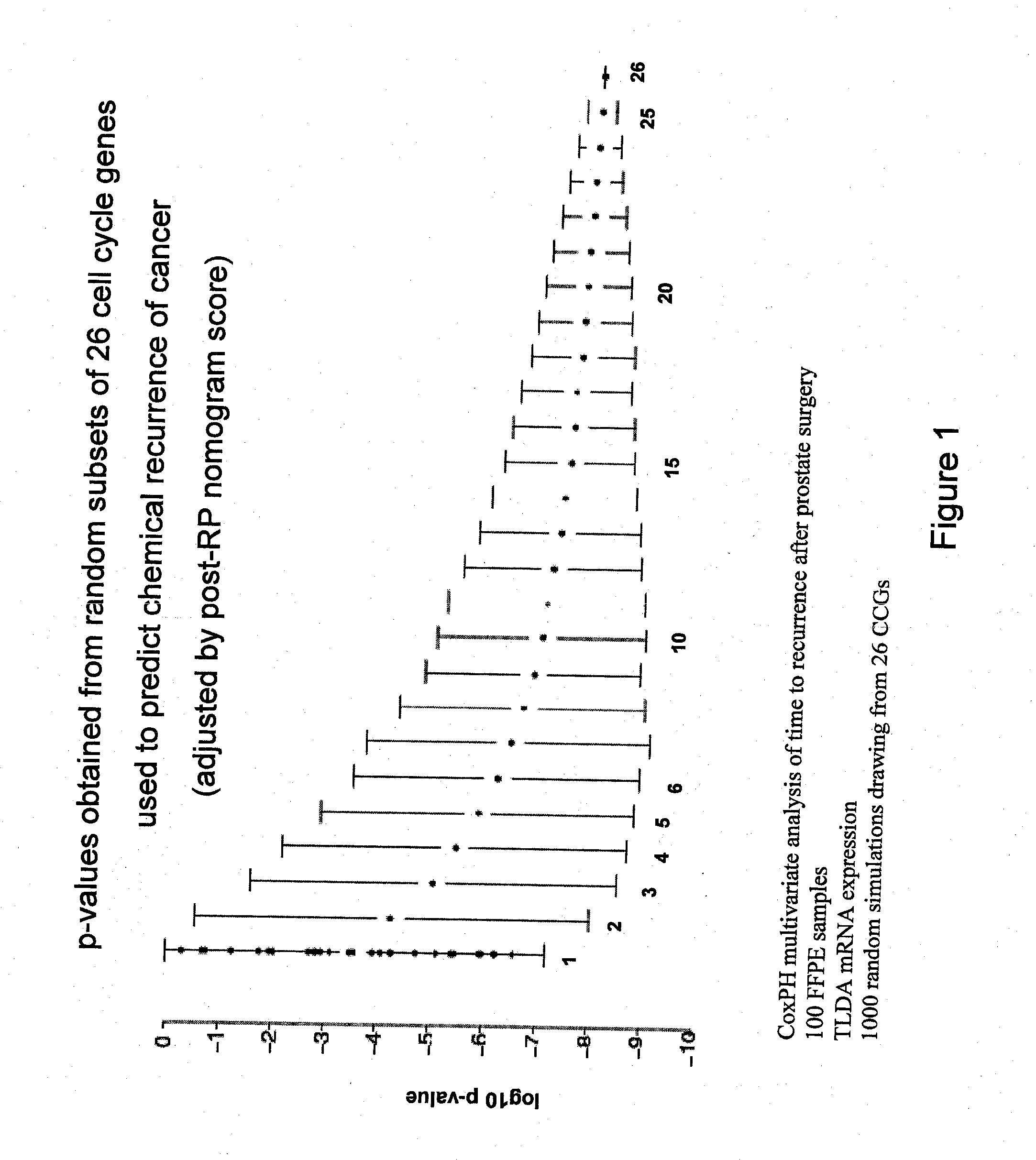
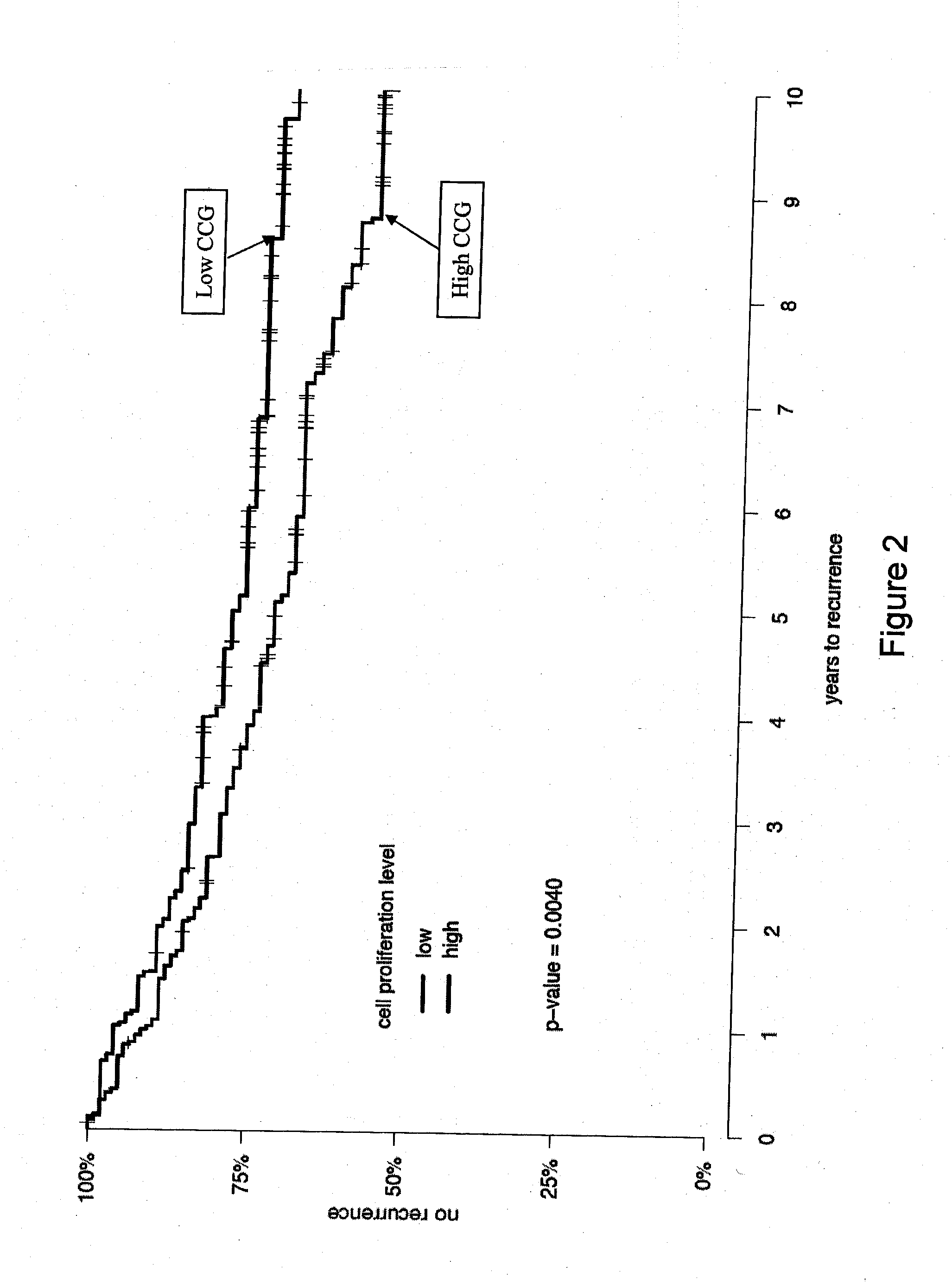
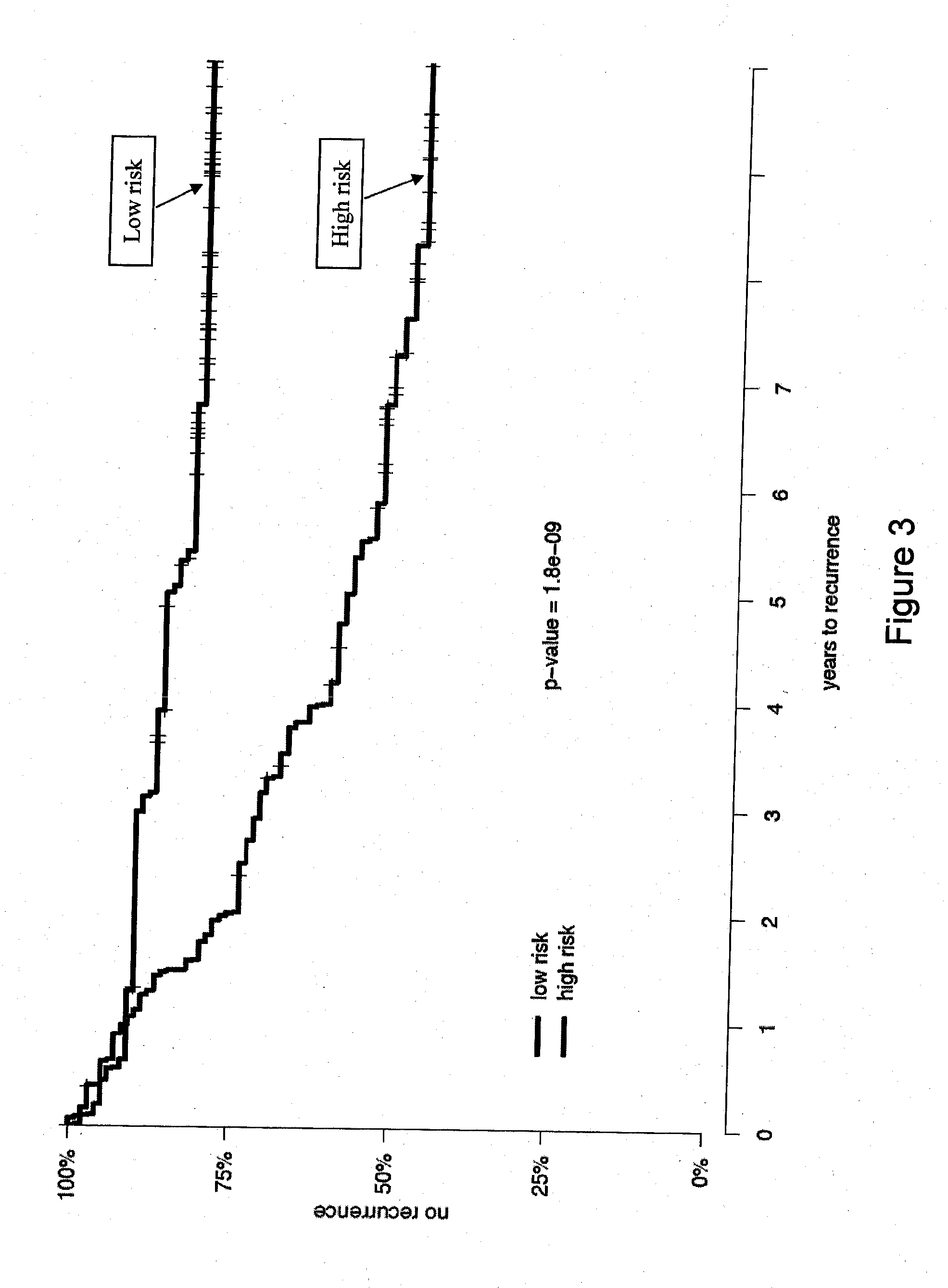
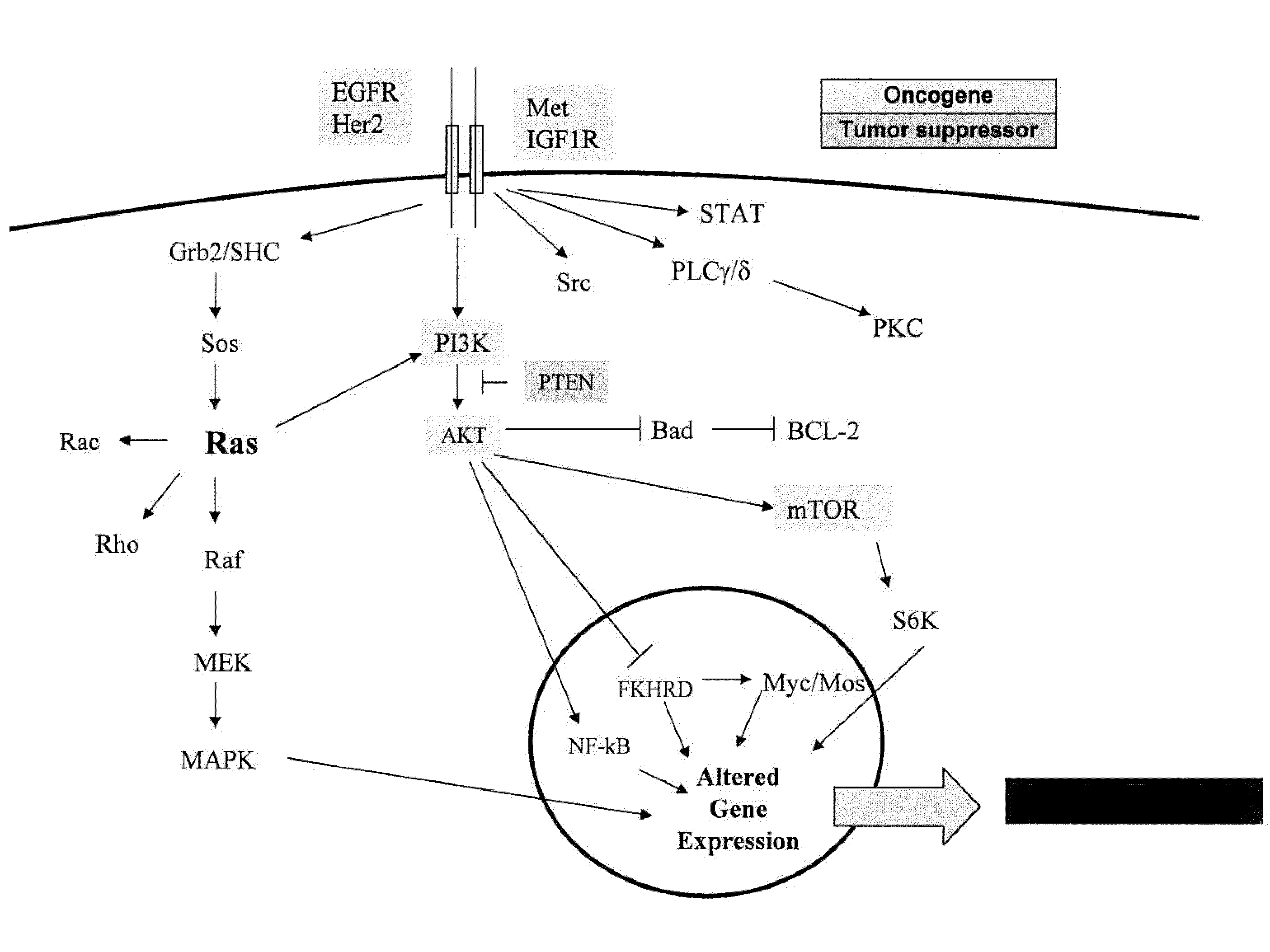
Patents
Literature
151 results about "Gene signature" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
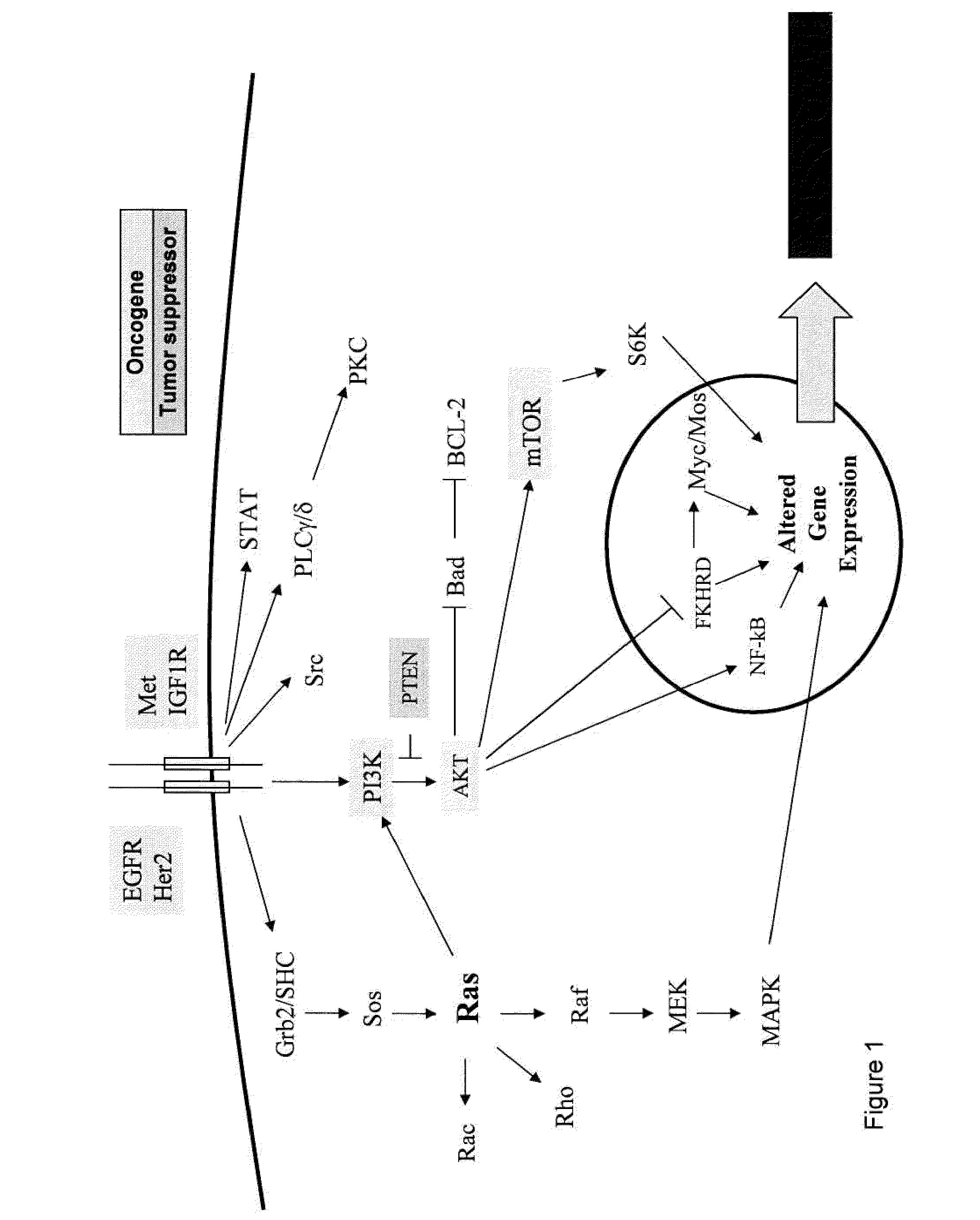
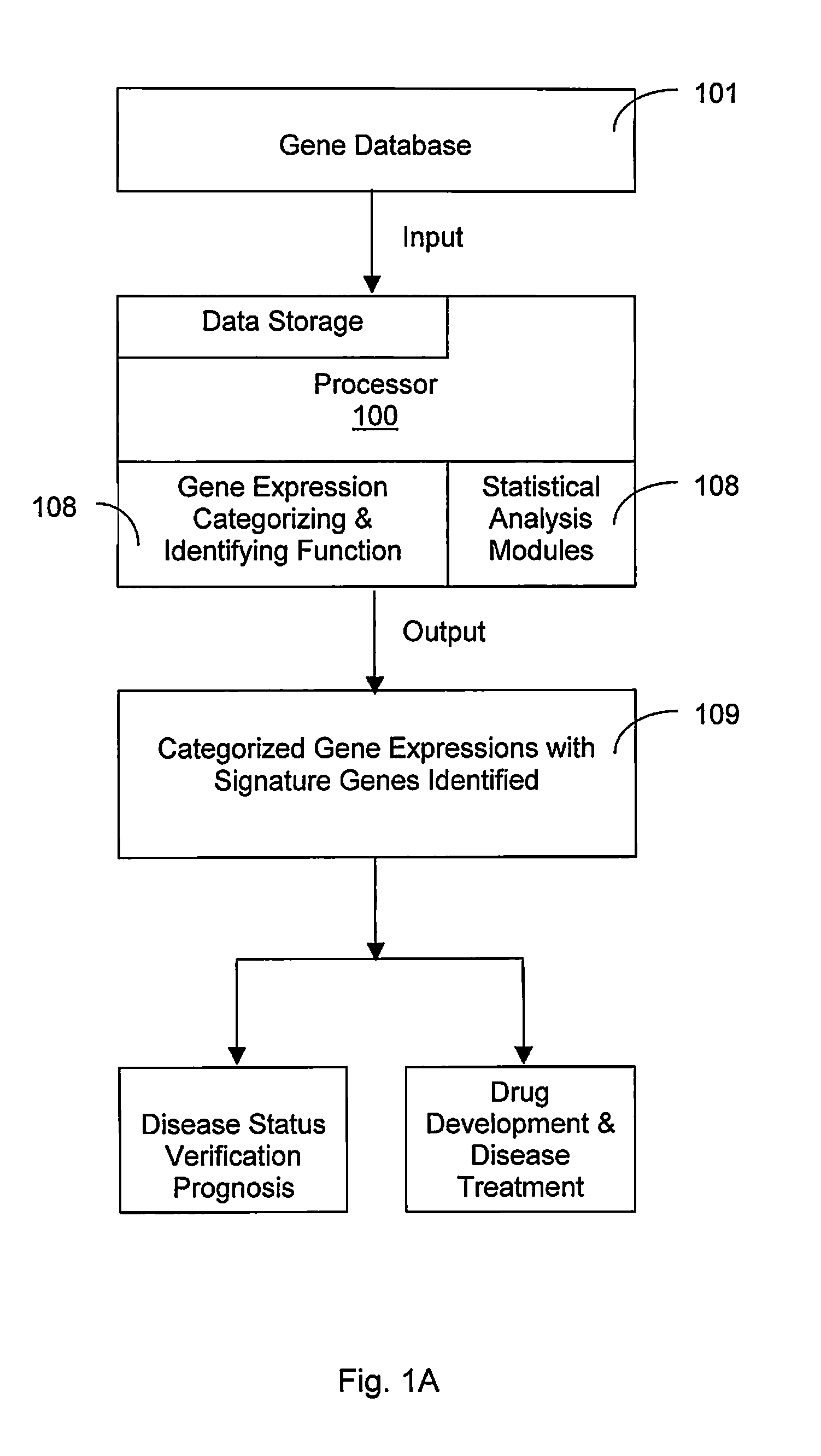
A gene signature or gene expression signature is a single or combined group of genes in a cell with a uniquely characteristic pattern of gene expression that occurs as a result of an altered or unaltered biological process or pathogenic medical condition. This is not to be confused with the concept of gene expression profiling. Activating pathways in a regular physiological process or a physiological response to a stimulus results in a cascade of signal transduction and interactions that elicit altered levels of gene expression, which is classified as the gene signature of that physiological process or response. The clinical applications of gene signatures breakdown into prognostic, diagnostic and predictive signatures. The phenotypes that may theoretically be defined by a gene expression signature range from those that predict the survival or prognosis of an individual with a disease, those that are used to differentiate between different subtypes of a disease, to those that predict activation of a particular pathway. Ideally, gene signatures can be used to select a group of patients for whom a particular treatment will be effective.
Compositions and methods for treating and diagnosing cancer
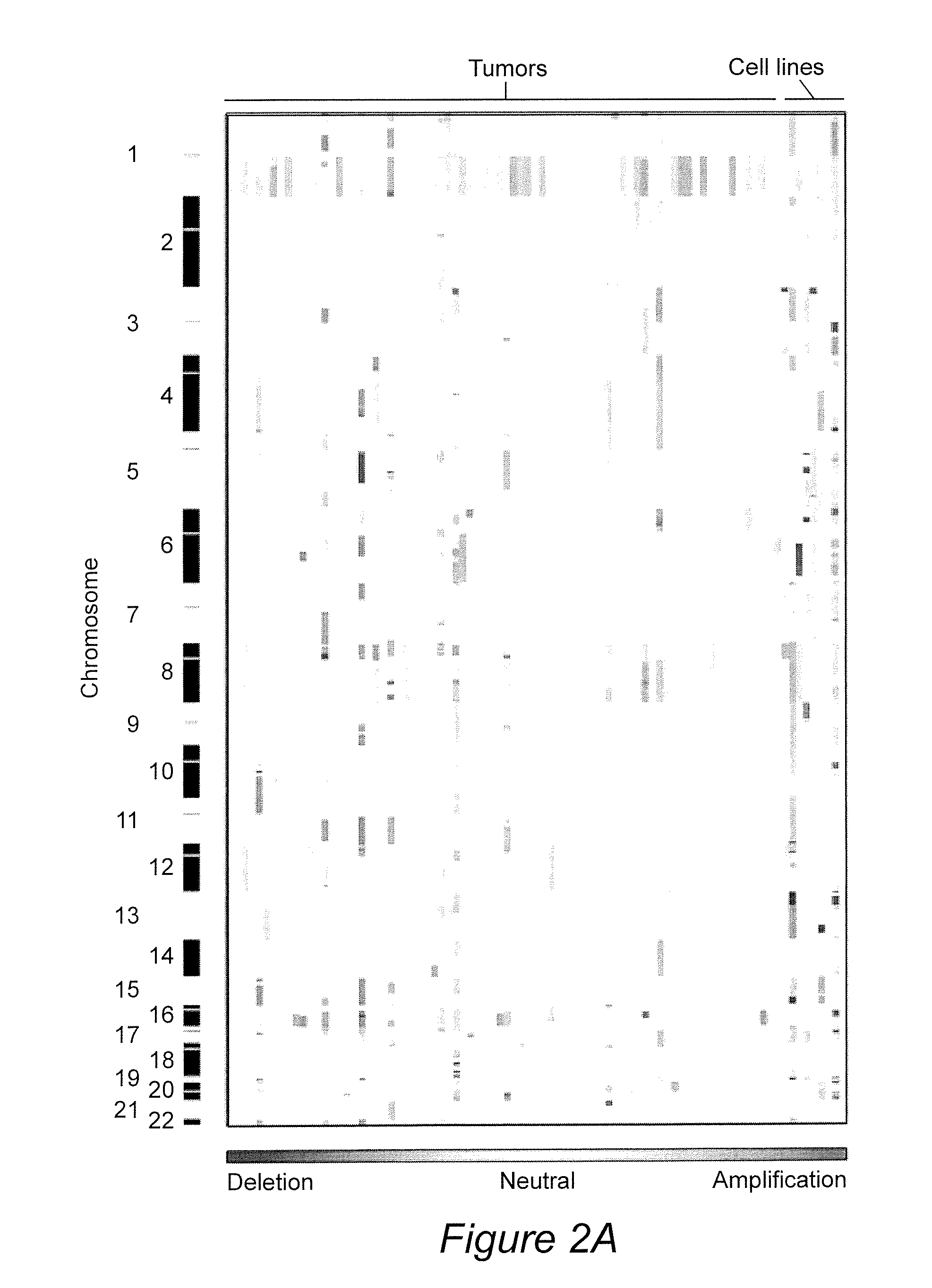
InactiveUS20070105133A1High of deathHigh riskMicrobiological testing/measurementBiological material analysisCancer metastasisSolid tumor
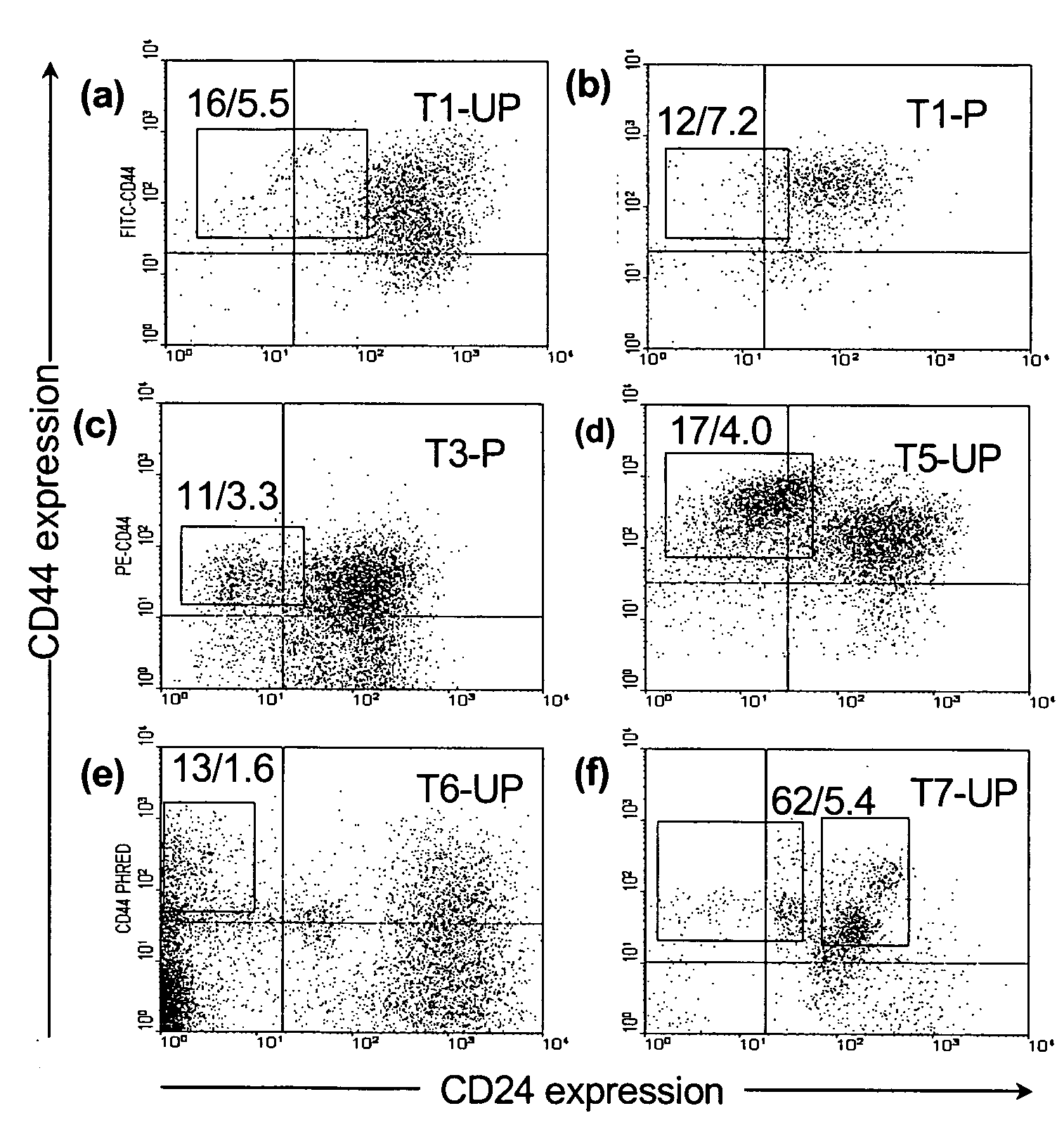
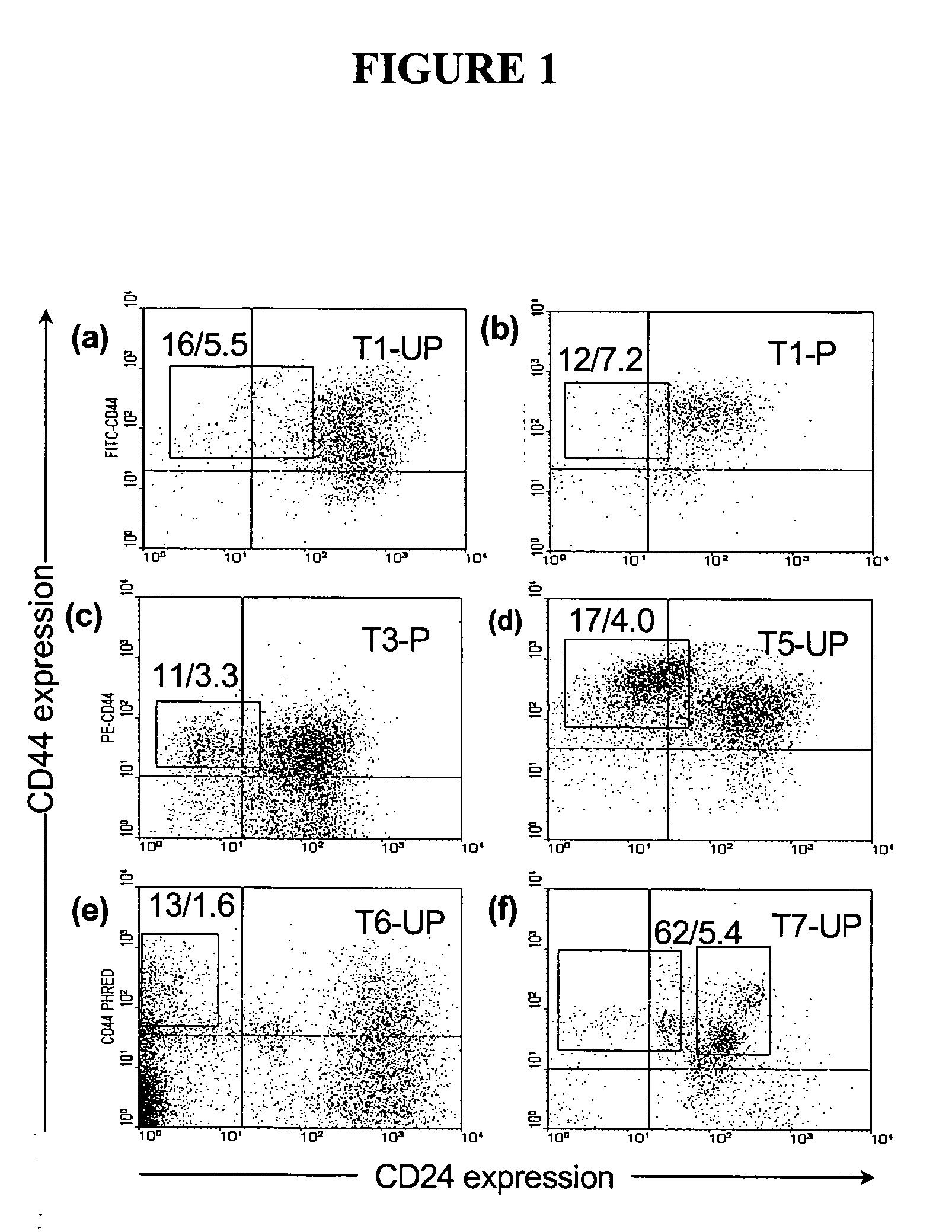
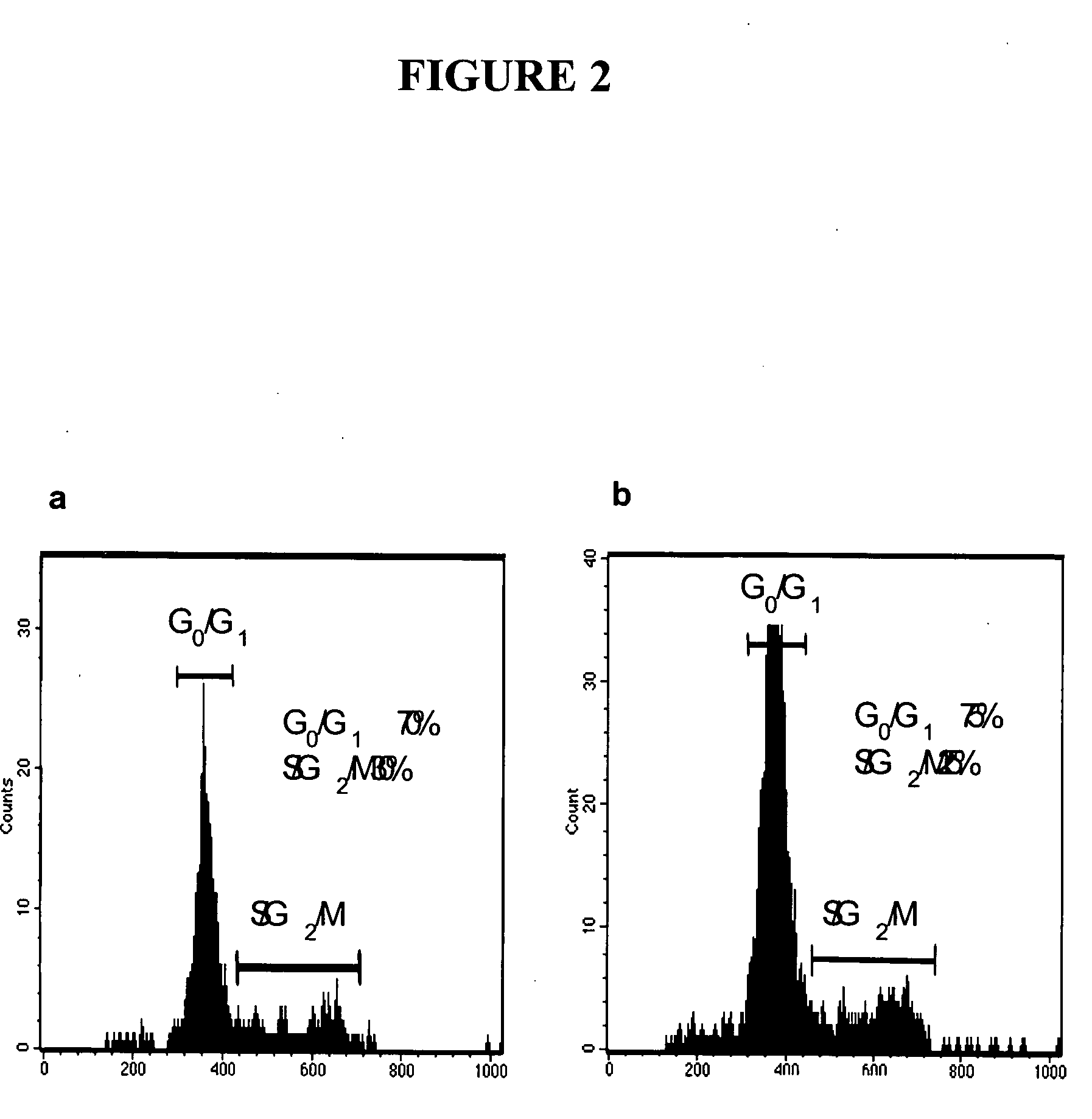
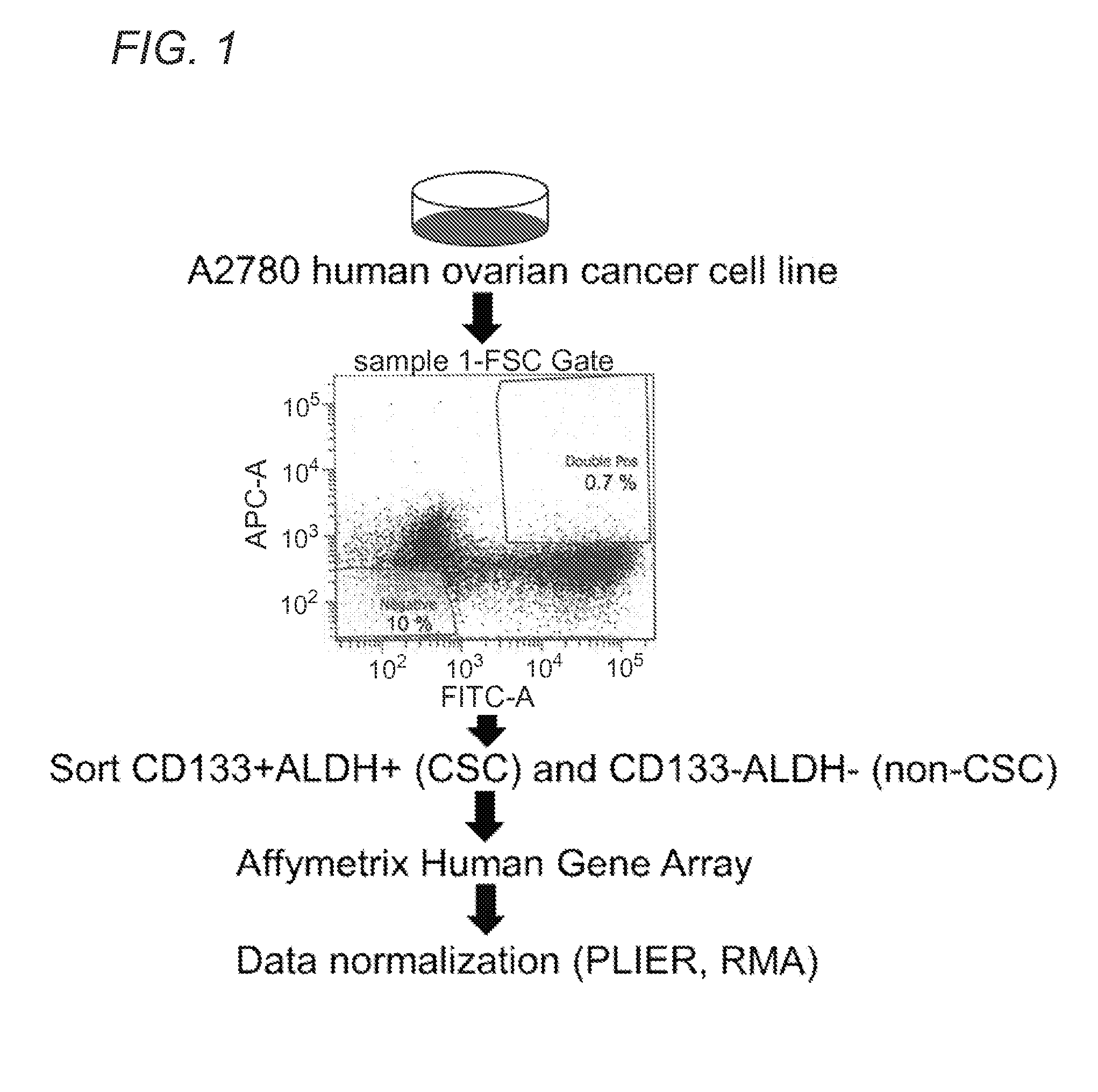
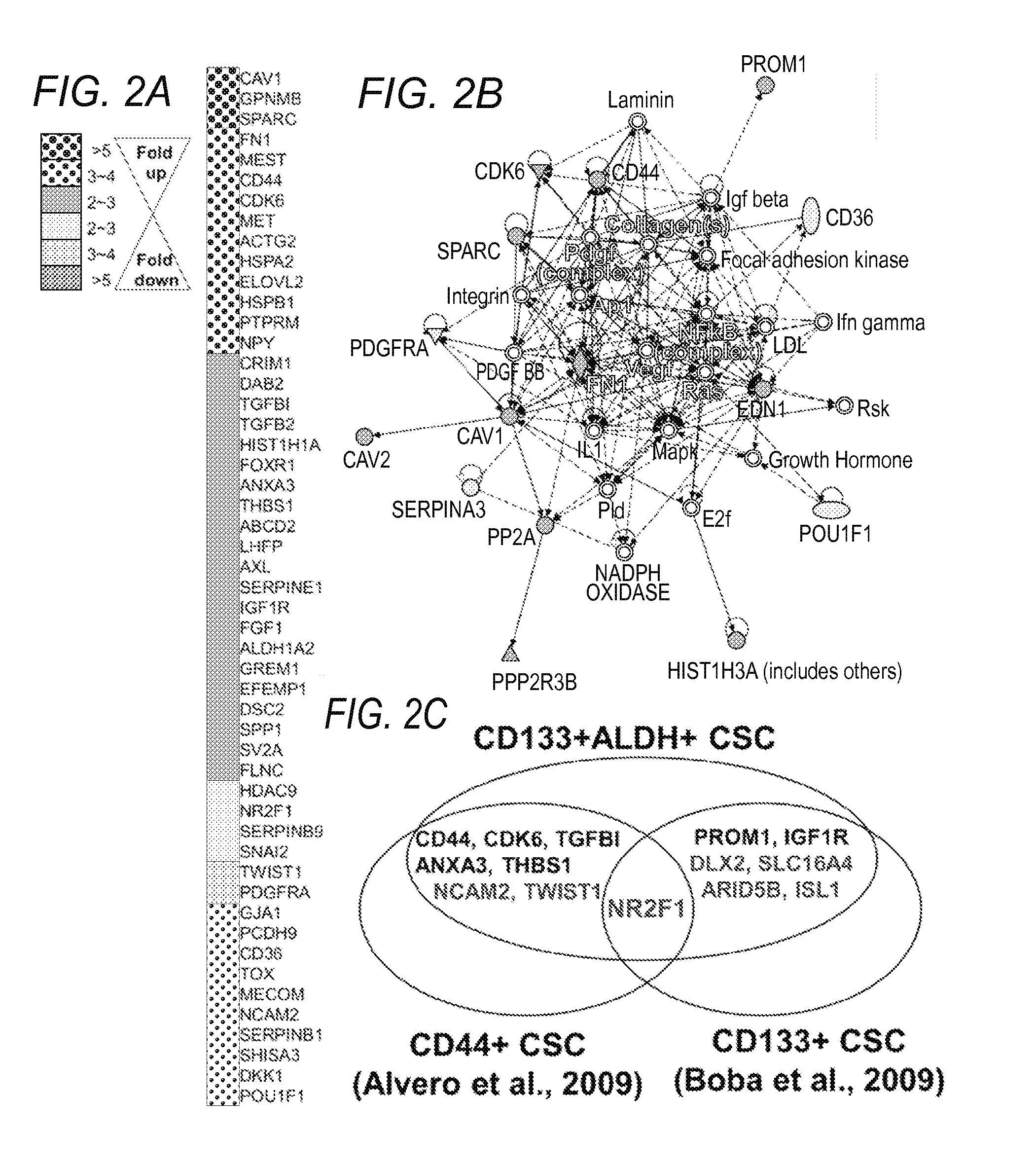
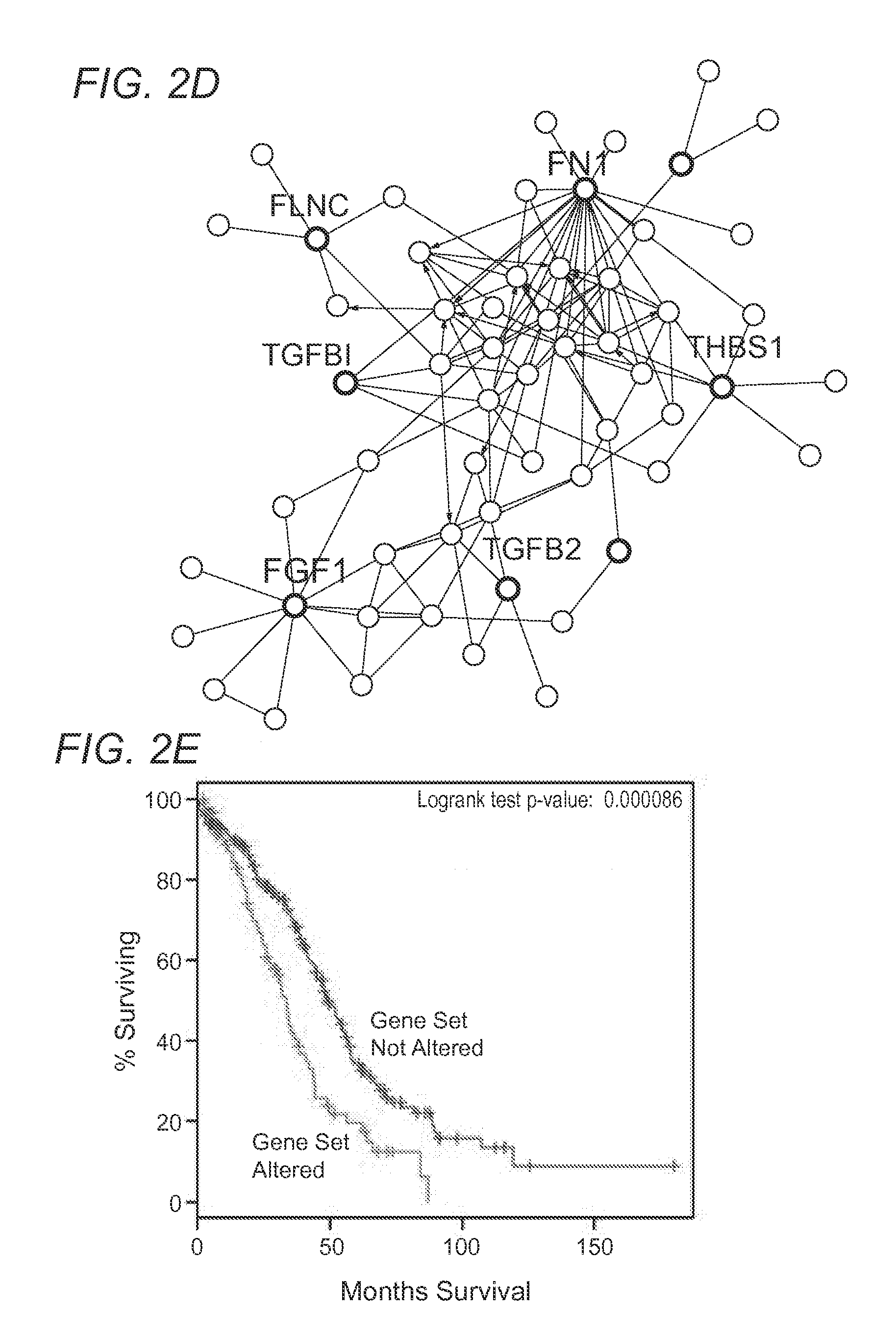
The present invention relates to compositions and methods for treating, characterizing, and diagnosing cancer. In particular, the present invention provides gene expression profiles and signatures associated with solid tumor stem cells, as well as novel stem cell cancer markers useful for the diagnosis, characterization, prognosis and treatment of solid tumor stem cells. More particularly, the present invention identifies two profiles of cancer stem cells useful for the diagnosis, characterization, and treatment of cancer and cancer metastases. The invention also provides a variety of reagents such as stem cell gene signatures for use in the diagnosis and management of cancer.
Owner:RGT UNIV OF MICHIGAN
Systems and methods for diagnosing & treating psychological and behavioral conditions
InactiveUS20050084880A1Bioreactor/fermenter combinationsBiological substance pretreatmentsGene signatureGene
The systems and methods described herein include microarray systems and methods for manufacturing and printing microarrays to provide gene chips capable of detecting signatures of psychiatric conditions, and as well as gene chips and arrays of sequences for such applications. The invention further provides methods of identifying gene signatures for psychiatric conditions, methods of treating such conditions, and methods of identifying therapeutics for the treatment of neurological and psychiatric conditions.
Owner:YALE UNIV
Colon cancer gene expression signatures and methods of use
InactiveUS10196691B2Accurate diagnosisQuality improvementMicrobiological testing/measurementBiological testingCancer genesOncology
A gene expression signature of colon cancer, microarrays including them and methods of using the colon gene expression signature are provided. The gene expression signature is especially useful for determining the prognosis of a patient diagnosed with colon cancer, such as stage II colon cancer. The gene signature described herein is also useful for determining effectiveness of surgical resection with or without adjuvant chemotherapy, and determining possibility of cancer recurrence in patients with colon cancer.
Owner:ALMAC DIAGNOSTICS SERVICES LIMITED
Use of emt gene signatures in cancer drug discovery, diagnostics, and treatment
The present invention provides diagnostic methods for assessing the EMT status of tumor cells, and for predicting the effectiveness of treatment of a cancer patient with an EGFR or IGF-1R kinase inhibitor, utilizing an EMT gene signature index score. The present invention further provides methods for treating patients with cancer that incorporate these methods.
Owner:OSI PHARMA LLC
Alzheimer's disease signature markers and methods of use
InactiveUS20140304845A1Sugar derivativesMicrobiological testing/measurementBiomarker (petroleum)Biology
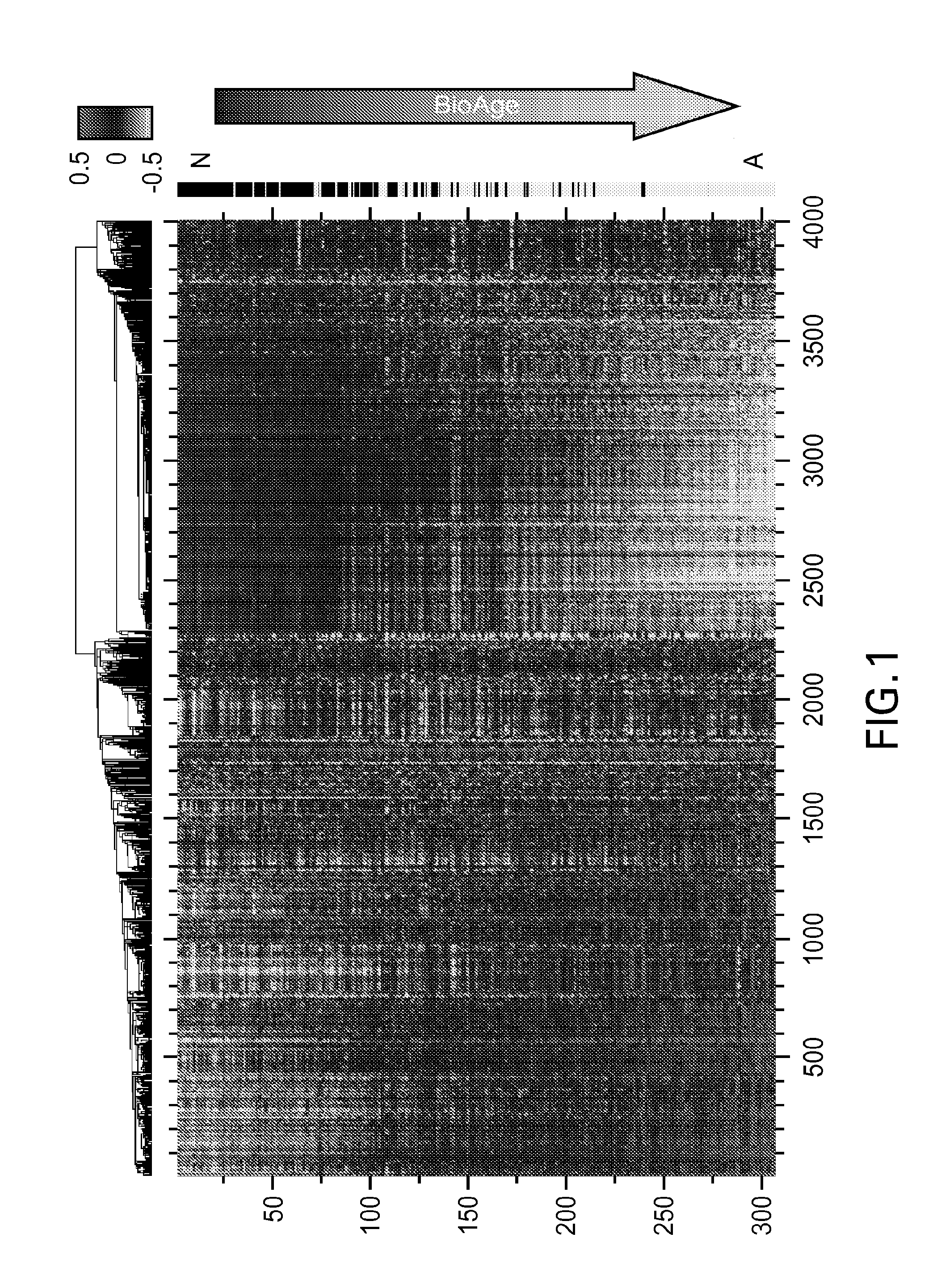
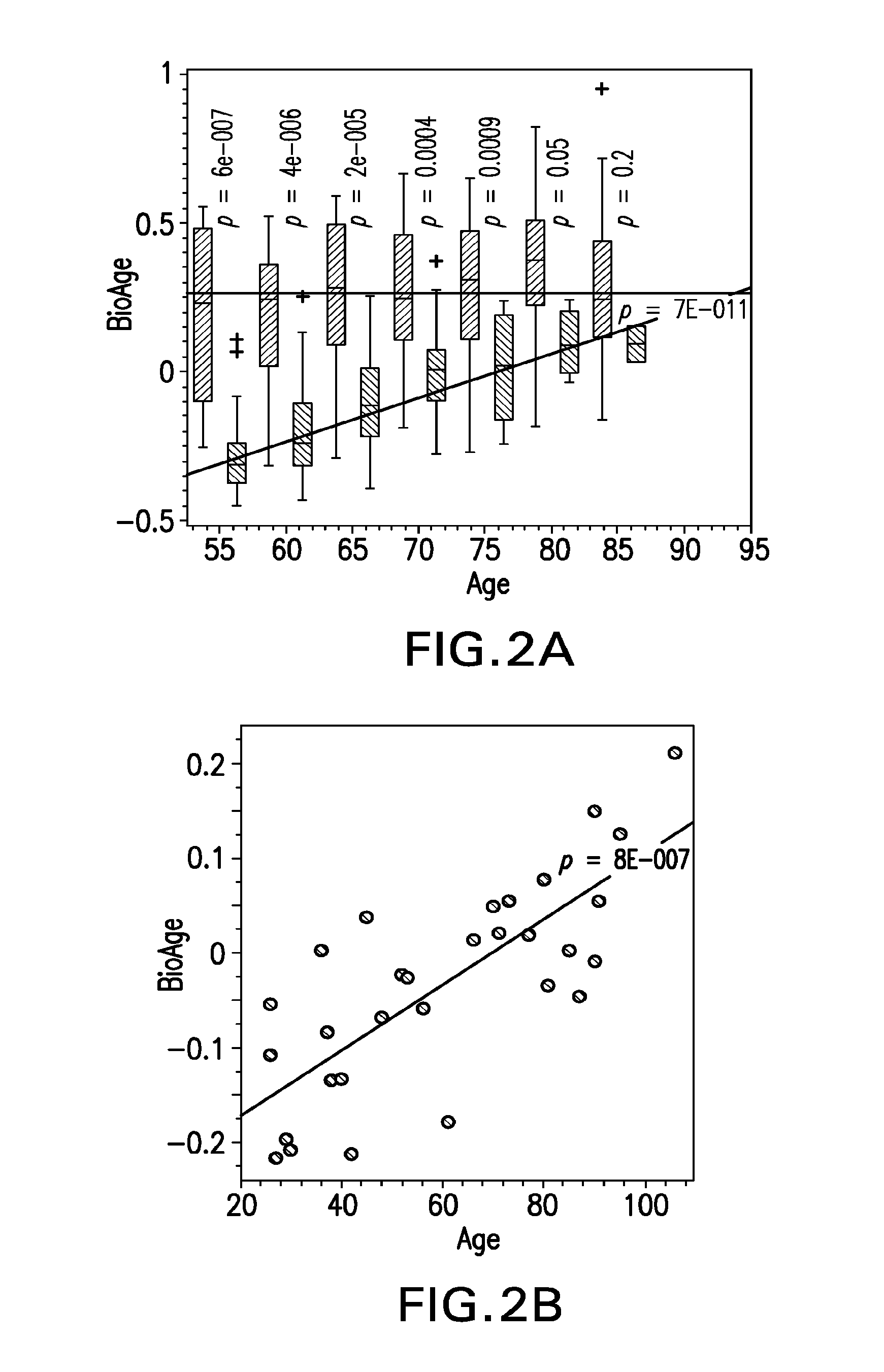
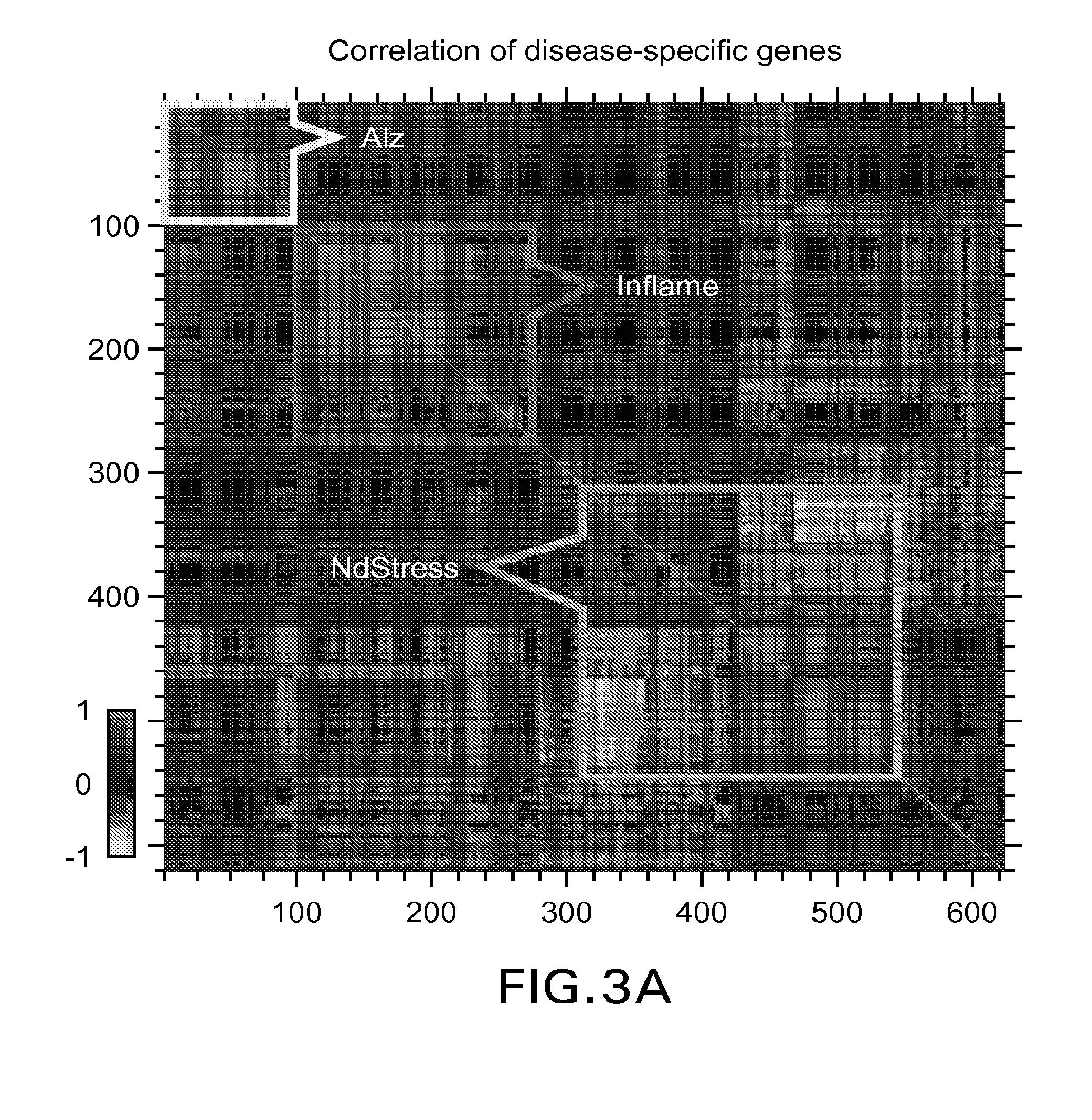
Methods, biomarkers, and expression signatures are disclosed for assessing the disease progression of Alzheimer's disease (AD). In one embodiment, BioAge (biological age), NdStress (neurodegenerative stress), Alz (Alzheimer), and Inflame (inflammation) are used as biomarkers of AD progression. In another aspect, the invention comprises a gene signature for evaluating disease progression. In still another embodiment, methods for evaluating disease progression are provided. In yet another embodiment, the invention can be used to identify animal models for use in the development and evaluation of therapeutics for the treatment of AD.
Owner:MERCK SHARP & DOHME CORP
Methods for predicting cancer outcome and gene signatures for use therein
InactiveUS20060195266A1Accurate predictionMicrobiological testing/measurementBiostatisticsSupport vector machineGene Feature
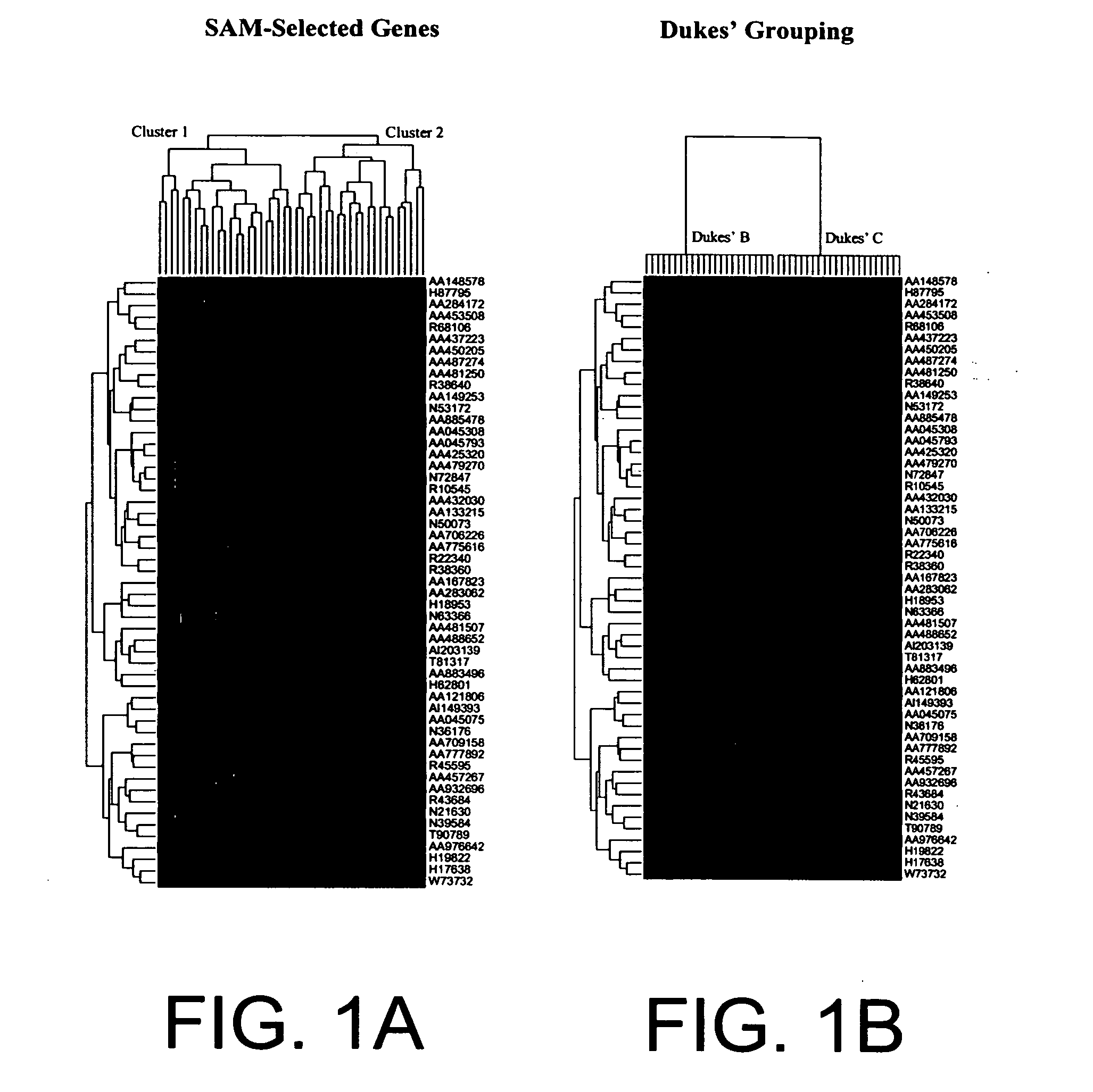
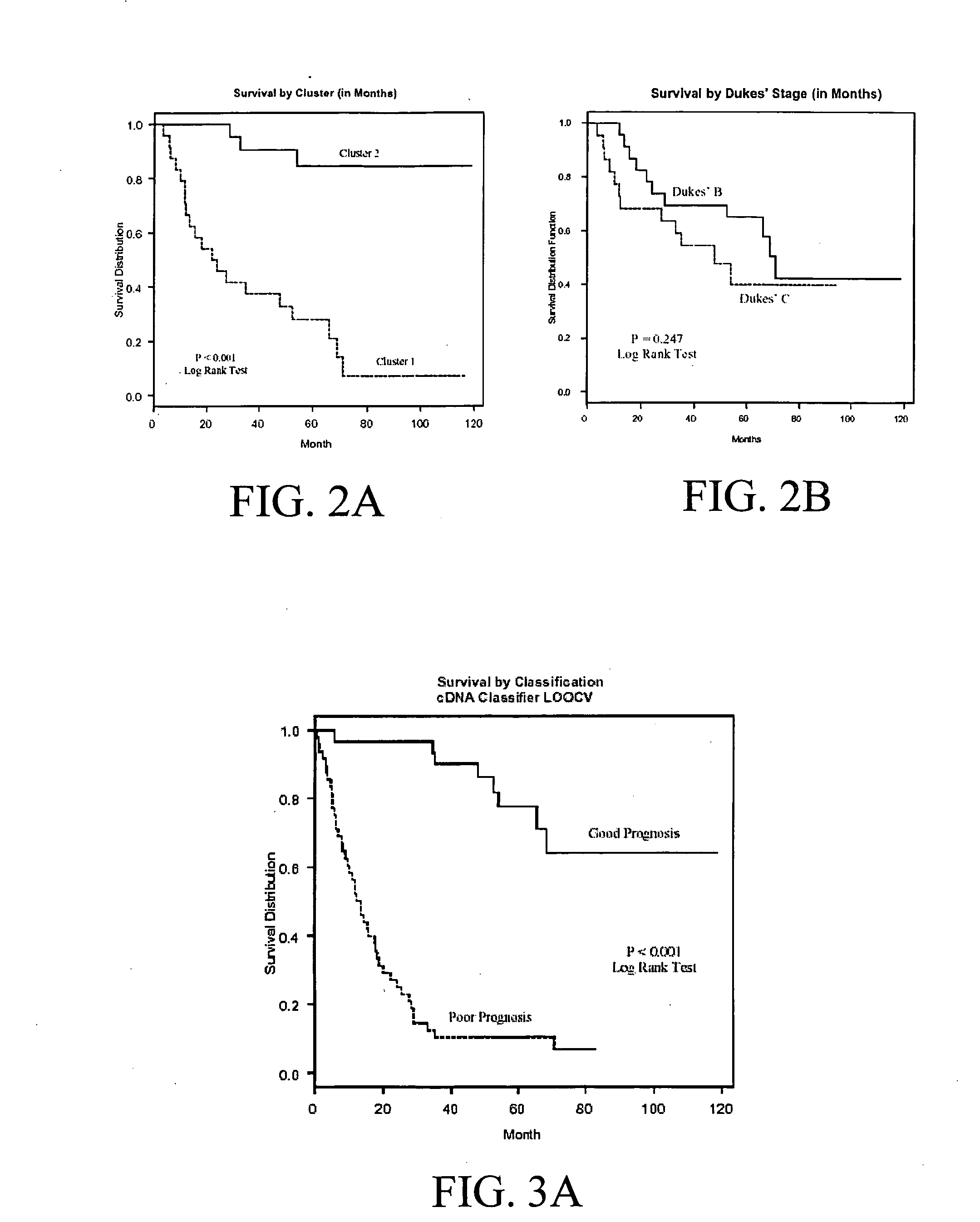
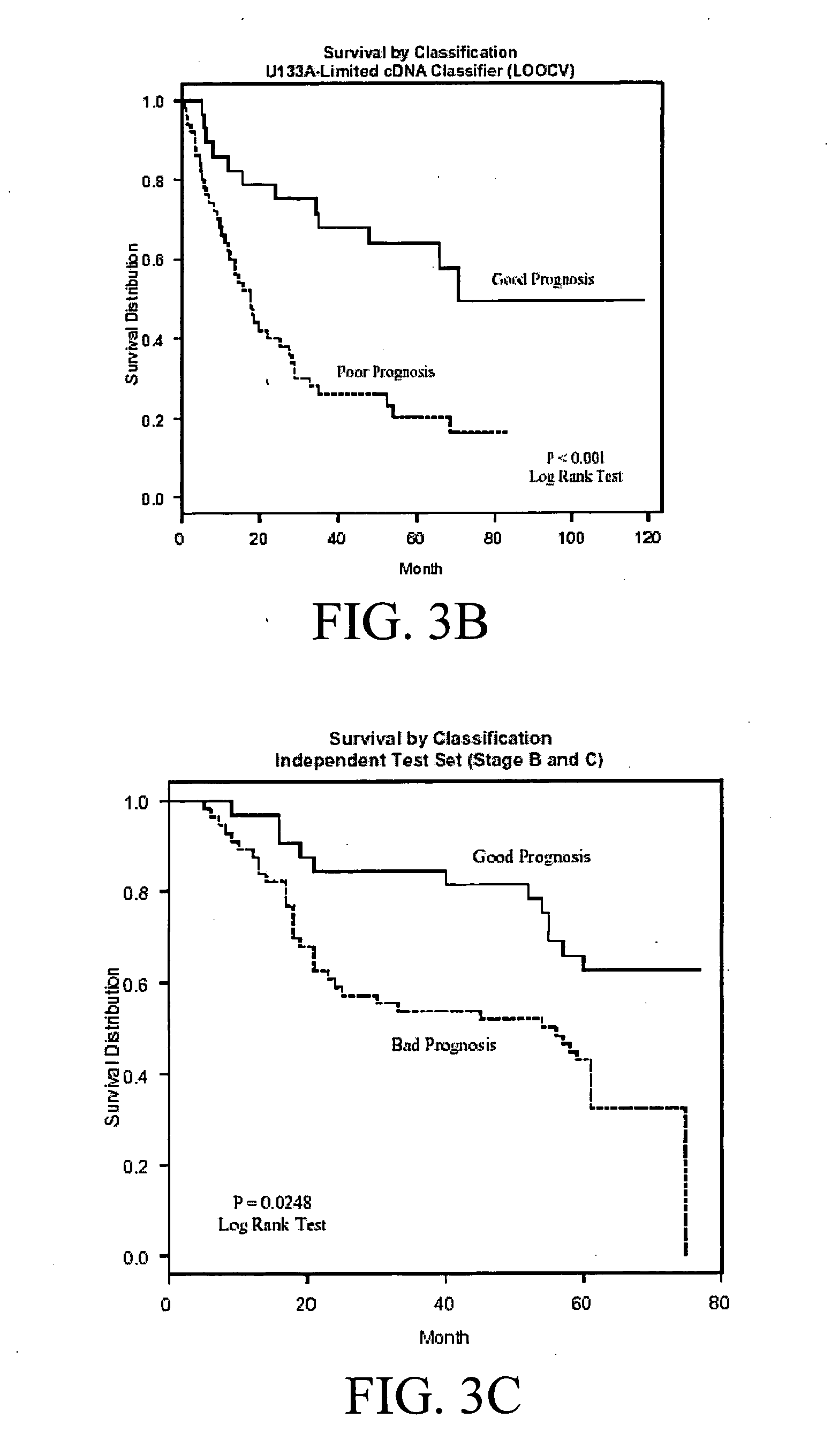
The present invention pertains to specific gene signatures for cancer that are used to predict survival and novel processes for identifying such gene signatures. In one embodiment, gene signatures for human colorectal cancer are identified and outcomes are linked to the specific gene signatures using significance analysis of microarrays (SAM) and support vector machines (SVM) to provide a prognosis / survival classifier.
Owner:YEATMAN TIMOTHY
Screening, Diagnosis and Prognosis of Autism and Other Developmental Disorders
ActiveUS20150227681A1Improve accuracyHigh sensitivityMicrobiological testing/measurementLibrary screeningDevelopmental disorderCrowds
The invention provides a method and system combining functional genomic and genetic, proteomic, anatomic neuroimaging, functional neuroimaging, behavioral and clinical measurements and data analyses for autism pediatric population screening, diagnosis or prognosis. More specifically, the invention provides a weighted gene and feature test for autism which uses a weighted gene signature matrix for comparison to a reference database of healthy and afflicted individuals. The invention also provides normalized gene expression value signatures for comparison to a reference database. The invention additionally combines either the weighted gene or the normalized gene analysis with comparisons to a gene-networks signature matrix, a multi-modal signature matrix, and a collateral features signature matrix for improved accuracy in screening, diagnostic and prognostic relevance for autism, particularly for newborns, babies ages birth to 1 year, toddlers ages 1 to 2 years, toddlers ages 2 to 3 years and young children ages 3 through 4 years.
Owner:RGT UNIV OF CALIFORNIA
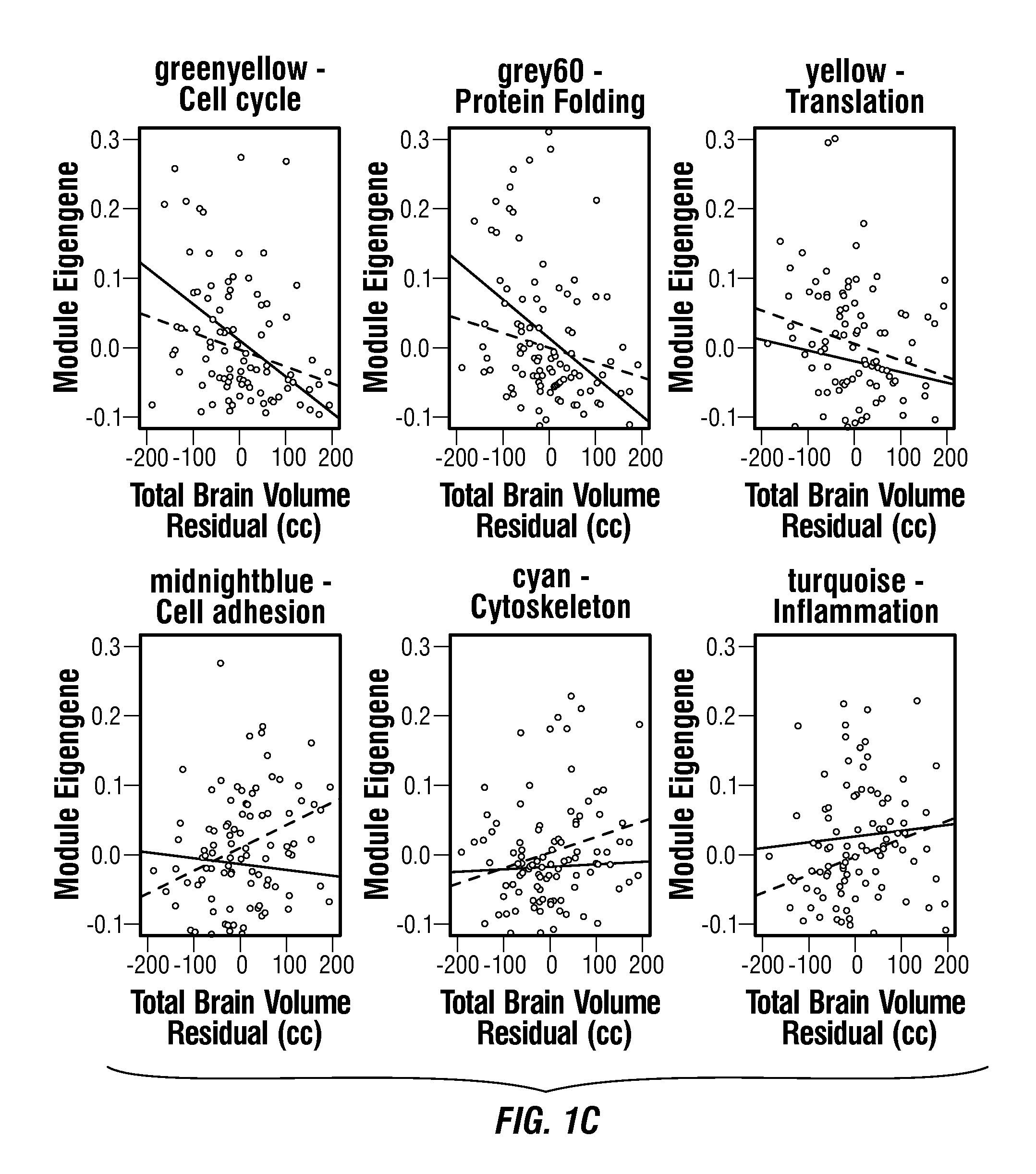
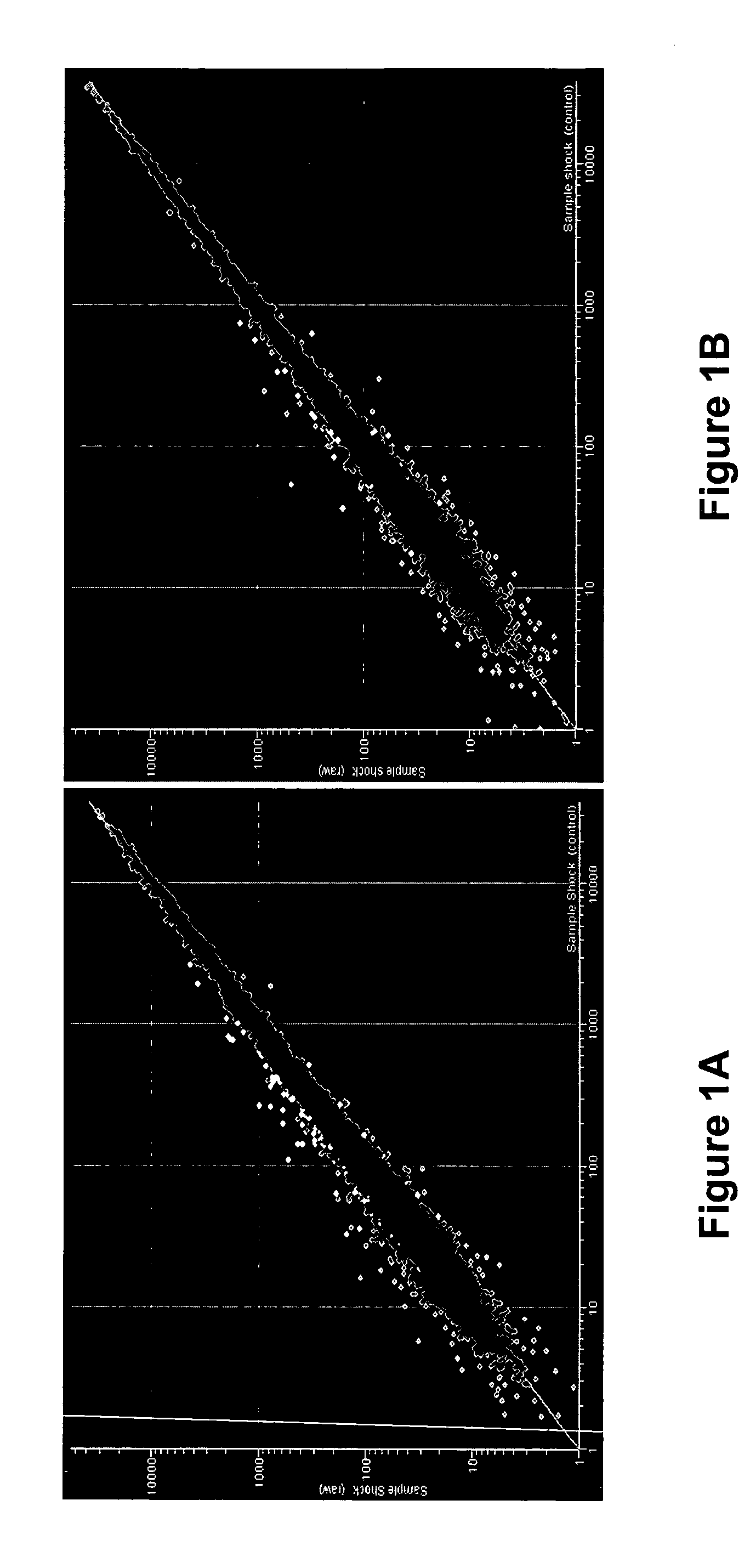
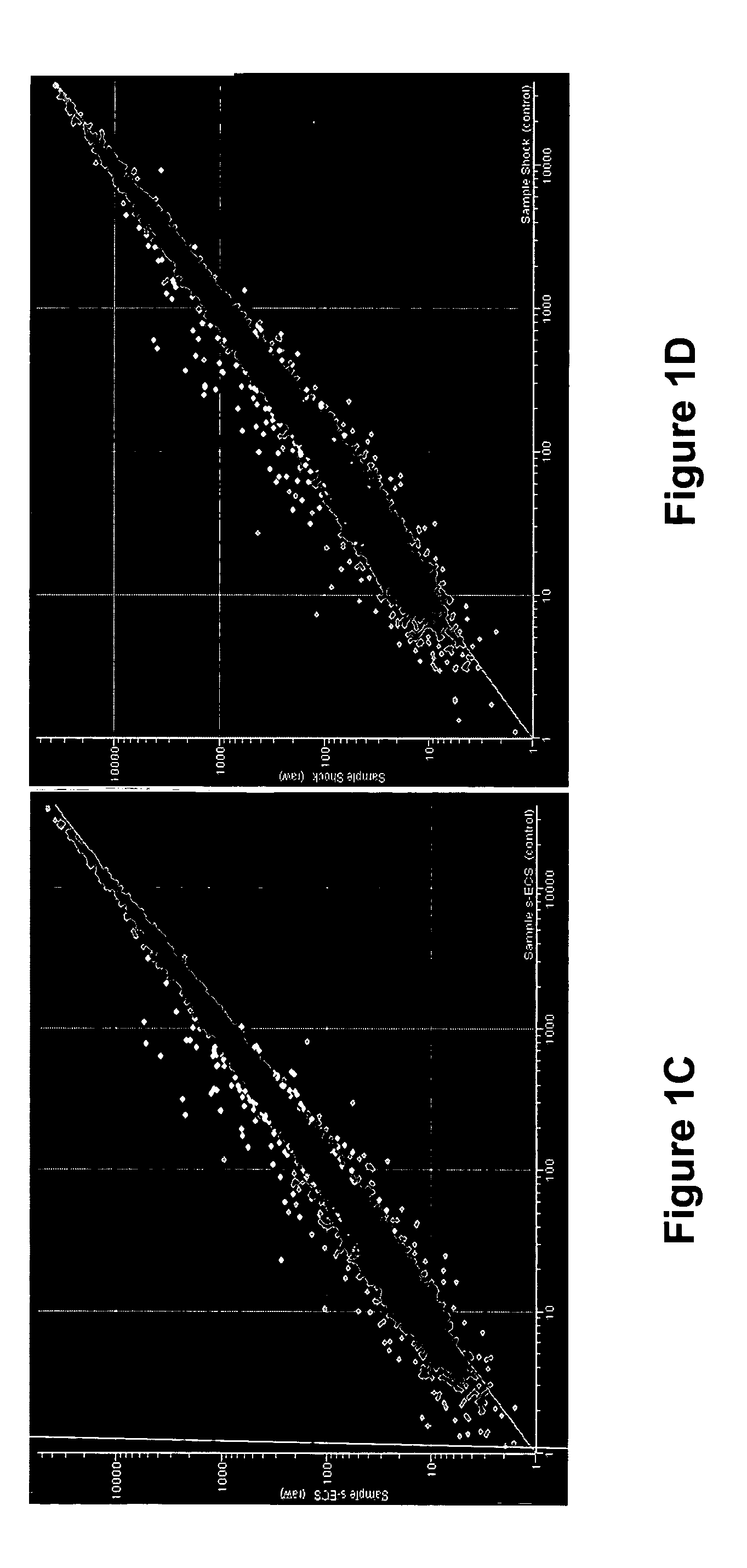
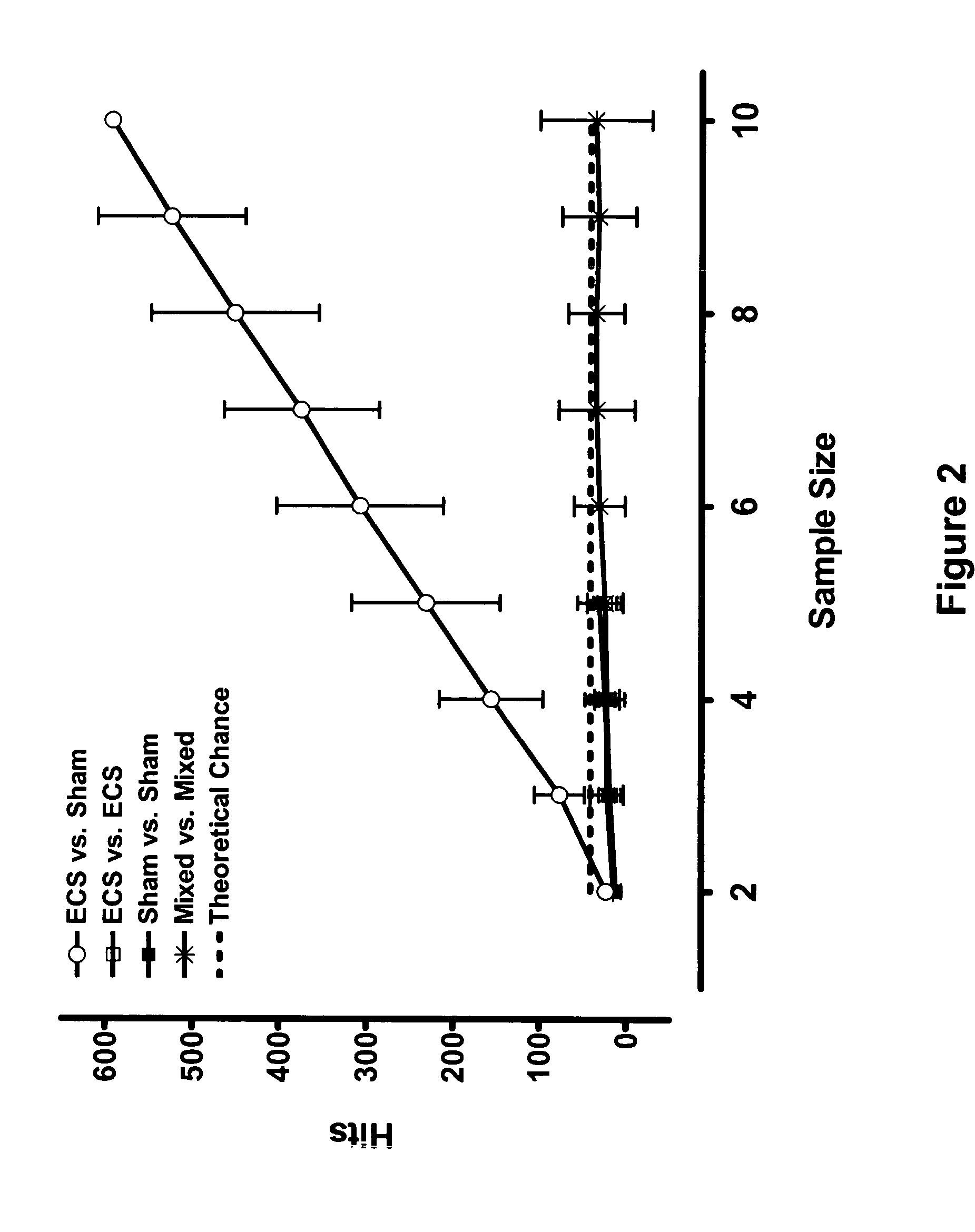
Gene signature of electroshock therapy and methods of use
InactiveUS20040152107A1Effective therapyCompound screeningApoptosis detectionDiseaseElectroconvulsive therapy
Gene signatures associated with electroconvulsive therapy (ECT) and / or electroconvulsive seizures (ECS) are disclosed. Such "signatures" are useful, e.g., for diagnosing and treating neuropsychiatric disorders such as major depressive disorder (MDD), bipolar affective disorder (BAD) and psychotic depression. Such methods are therefore also provided here. The invention additionally provides screening methods that use gene signatures of the invention to identify new therapeutic compounds for treating these disorders.
Owner:PSYCHIATRIC GENOMICS
Gene signatures for cancer prognosis
Biomarkers and methods using the biomarkers for the prediction of the recurrence risk of cancer in a patient are provided.
Owner:MYRIAD GENETICS
Methods and gene expression signature for assessing ras pathway activity
Methods, biomarkers, and expression signatures are disclosed for assessing the regulation status of RAS pathway signaling in a cell sample or subject. More specifically, several aspects of the invention provide a set of genes which can be used as biomarkers and gene signatures for evaluating RAS pathway deregulation status in a sample; classifying a cell sample as having a deregulated or regulated RAS signaling pathway; determining whether an agent modulates the RAS signaling pathway in sample; predicting response of a subject to an agent that modulates the RAS signaling pathway; assigning treatment to a subject; and evaluating the pharmacodynamic effects of cancer therapies designed to regulate RAS pathway signaling.
Owner:MERCK SHARP & DOHME CORP
Molecular signatures of ovarian cancer
ActiveUS20150322530A1Optimal cytoreductionHeavy metal active ingredientsOrganic active ingredientsTriageOncology
Described herein are gene signatures providing prognostic, diagnostic, treatment and molecular subtype classifications of ovarian cancers through generation of ovarian cancer disease signatures (OCDSs) that account for molecular heterogeneity present in gynecological cancers. An ovarian cancer fixed signature (OCFS) is described which relates to the core programming of disease development, in addition to an ovarian cancer stem cell (OCSC) signature. Development various disease signature, suggests personalized treatment strategies focused on molecular subtypes of gynecological cancers, such as triage tests for patients.
Owner:CEDARS SINAI MEDICAL CENT
Lung adenocarcinoma-related gene signatures and application thereof
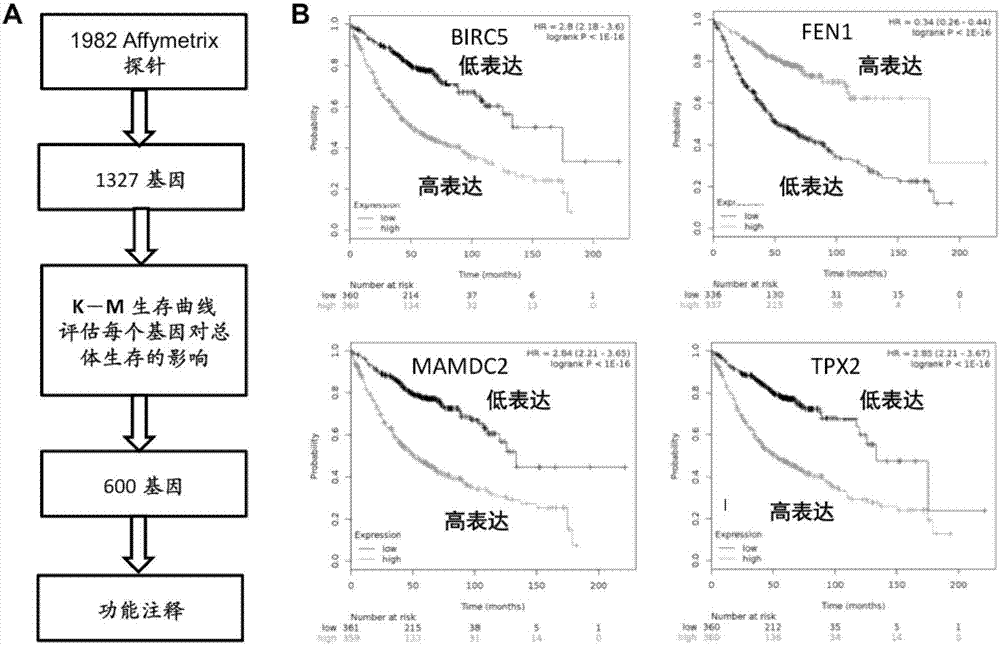
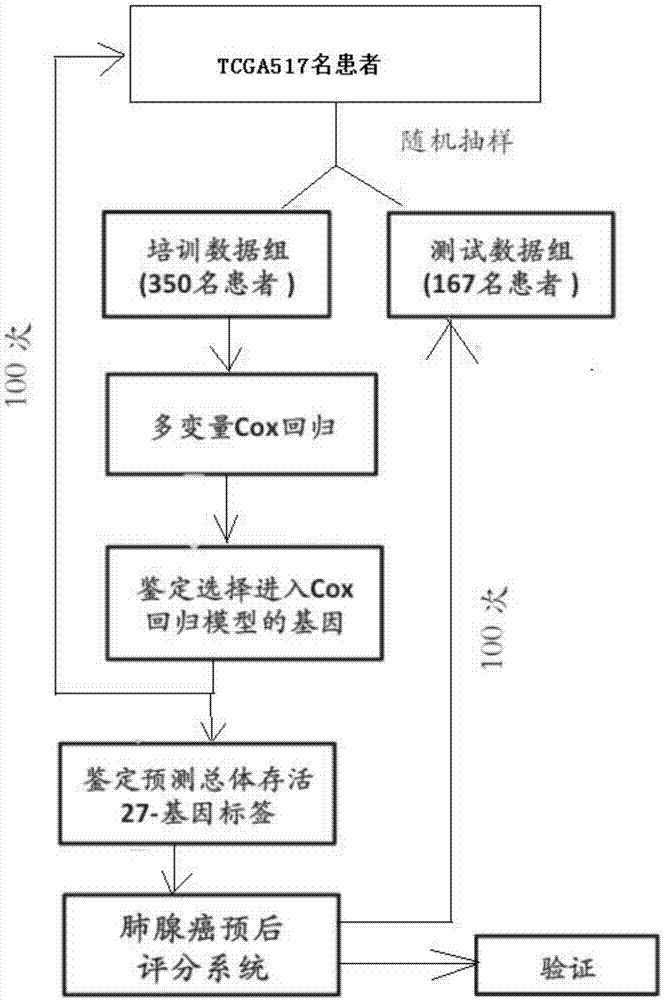
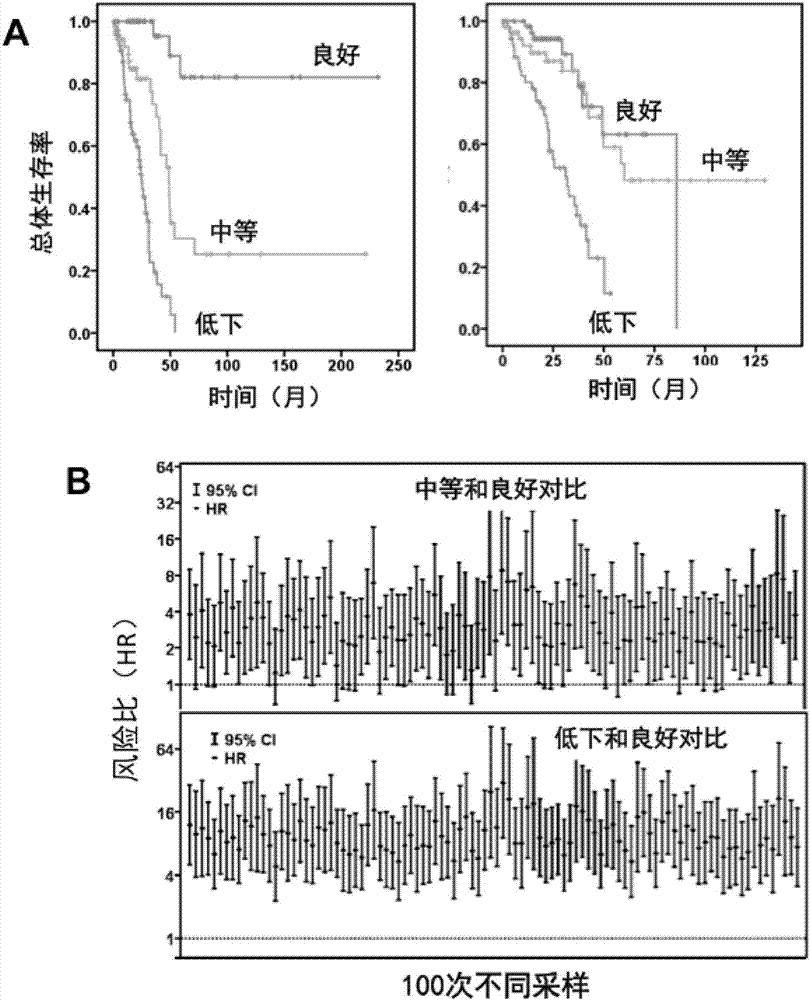
ActiveCN107881234AAvoid overdosingReduce medical costsMicrobiological testing/measurementAdenocarcinomaClinical prognosis
The invention relates to lung adenocarcinoma-related gene signatures and an application thereof. The invention relates to a group of 27 prognosis related gene signatures based on lung adenocarcinoma and a detection result of an expression level of the gene signatures in a clinical sample, a prediction value is calculated to evaluate clinical prognosis of lung adenocarcinoma, and the invention alsoprovides an application of the gene signatures. A system is used for preventing lung adenocarcinoma patient prognosis and guiding a clinical treatment decision, and a purpose of accurate or individualized medicine can be achieved. According to the system and different detection technical platforms, a corresponding 27 gene expression measuring kit is designed and exploited.
Owner:NANJING KDRB BIOTECH INC LTD
Compositions and methods for treating and diagnosing cancer
InactiveUS20100093556A1Microbiological testing/measurementLibrary screeningCancers diagnosisCancer metastasis
The present invention relates to compositions and methods for treating, characterizing, and diagnosing cancer. In particular, the present invention provides gene expression profiles and signatures associated with solid tumor stem cells, as well as novel stem cell cancer markers useful for the diagnosis, characterization, prognosis and treatment of solid tumor stem cells. More particularly, the present invention identifies two profiles of cancer stem cells useful for the diagnosis, characterization, and treatment of cancer and cancer metastases. The invention also provides a variety of reagents such as stem cell gene signatures for use in the diagnosis and management of cancer.
Owner:ONCOMED PHARMA INC +1
Combined diagnosis marker for predicting small hepatocellular carcinoma relapse
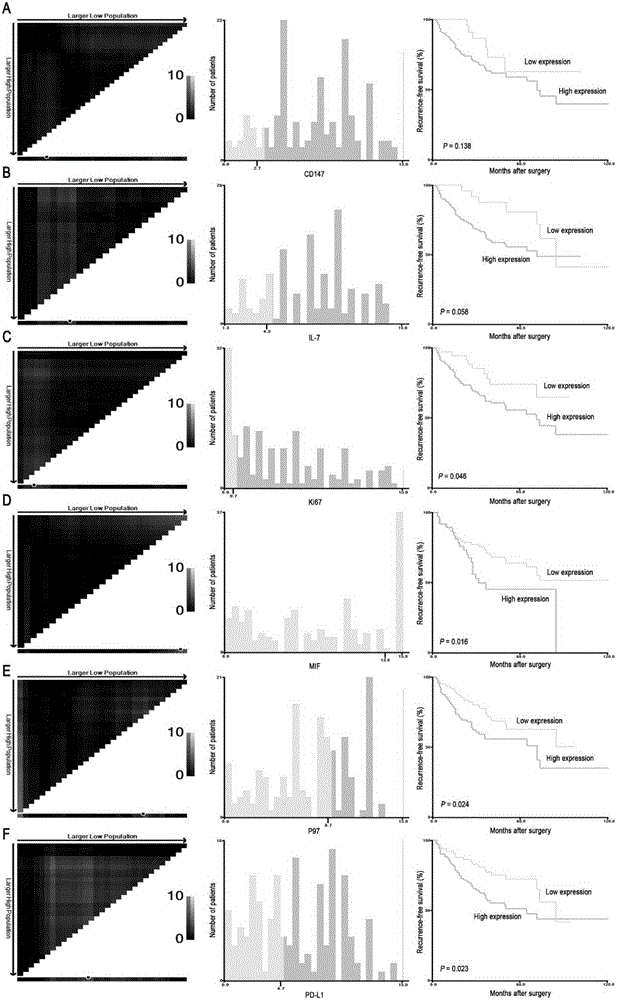
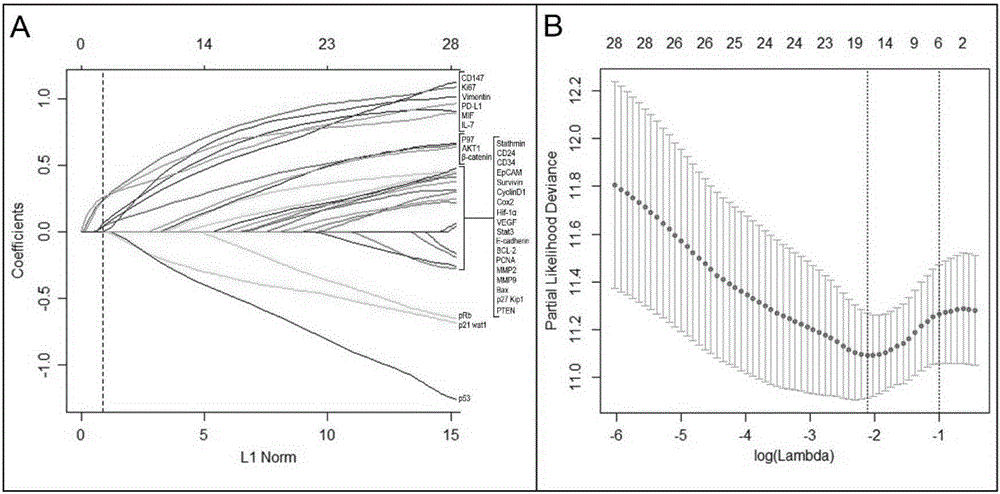
ActiveCN105319364AImprove forecast accuracyIncreased prognostic predictive valueMaterial analysisClinical valueIndividualized treatment
The invention discloses a combined diagnosis marker for predicting small hepatocellular carcinoma relapse. Based on an LASSO Cox regression model, the inventor confirms and establishes a small hepatocellular carcinoma relapse prediction model based on 6 gene signatures (including CD147, IL-7, Ki67, MIF, P97 and PD-L1) through analyzing a large number of cases; the risk ratio, defined by a 6 gene signature classifier, of a high relapse risk crowd to a low relapse risk crowd is 2.721 (95% CI: 1.633-4.532; P is smaller than 0.0001); the analysis of an internal validation group and an external validation group has the same result. The experiment proves that the classifier based on the 6 gene signatures can effectively predict different relapse risks of patients suffering from small hepatocellular carcinoma, the traditional diagnosis accuracy rate, predicted through clinicopathological parameters, of small hepatocellular carcinoma relapse is increased, the relapse risk of AFP-negative patients suffering from small hepatocellular carcinoma can be well judged, and the combined diagnosis marker has potential clinical value in the individualized treatment of small hepatocellular carcinoma.
Owner:SUN YAT SEN UNIV CANCER CENT
Pro-angiogenic genes in ovarian tumor endothelial cell isolates
InactiveUS20100286237A1Reduce and eliminate ovarian cancerDevelopment of therapyOrganic active ingredientsMicrobiological testing/measurementCell fractionationCell tumor
A gene profiling signature for ovarian tumor endothelial cells is disclosed herein. The gene signature can be used to diagnosis or prognosis an ovarian tumor, identify agents to treat an ovarian tumor, to predict the metastatic potential of an ovarian tumor and to determine the effectiveness of ovarian tumor treatments. Thus, methods are provided for identifying agents that can be used to treat ovarian cancer, for determining the effectiveness of an ovarian tumor treatment, or to diagnose or prognose an ovarian tumor. Methods of treatment are also disclosed which include administering a composition that includes a specific binding agent that specifically binds to one of the disclosed ovarian endothelial cell tumor-associated molecules and inhibits ovarian tumor in the subject.
Owner:US DEPT OF HEALTH & HUMAN SERVICES +1
Sense-antisense gene pairs for patient stratification, prognosis, and therapeutic biomarkers identification
InactiveUS20160259883A1Quality improvementHighly prognostically significantMicrobiological testing/measurementLibrary screeningPrognostic signaturePatient stratification
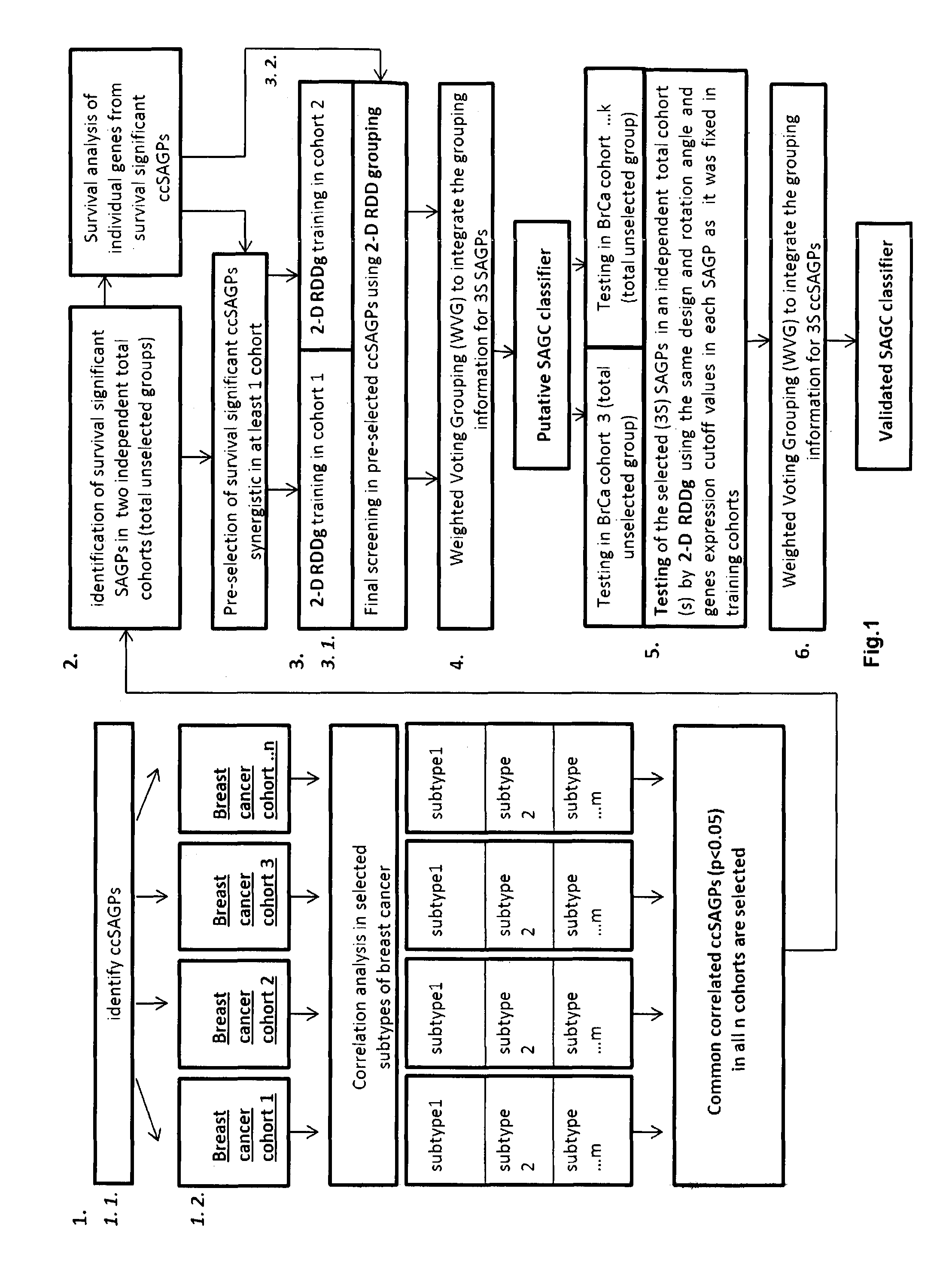
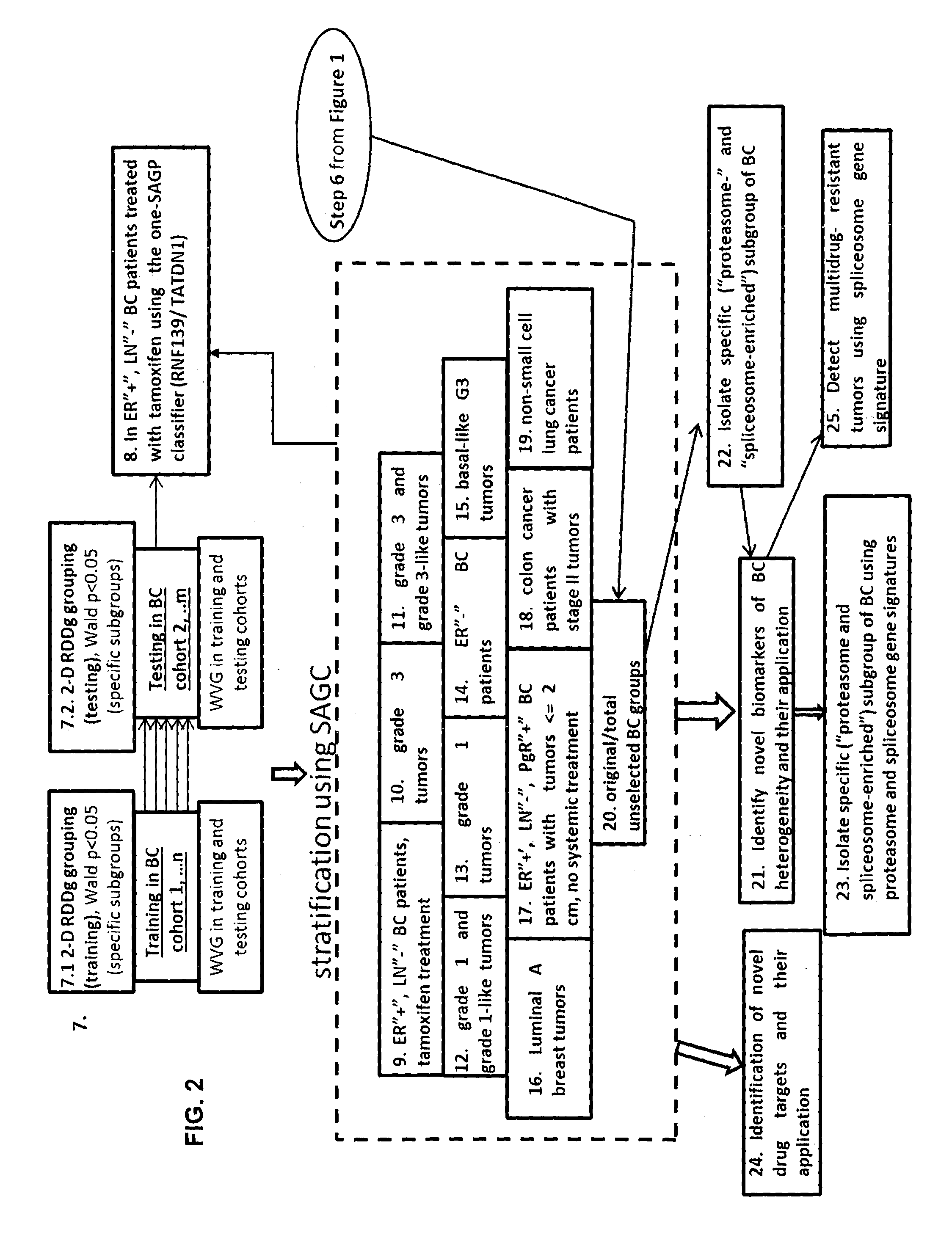
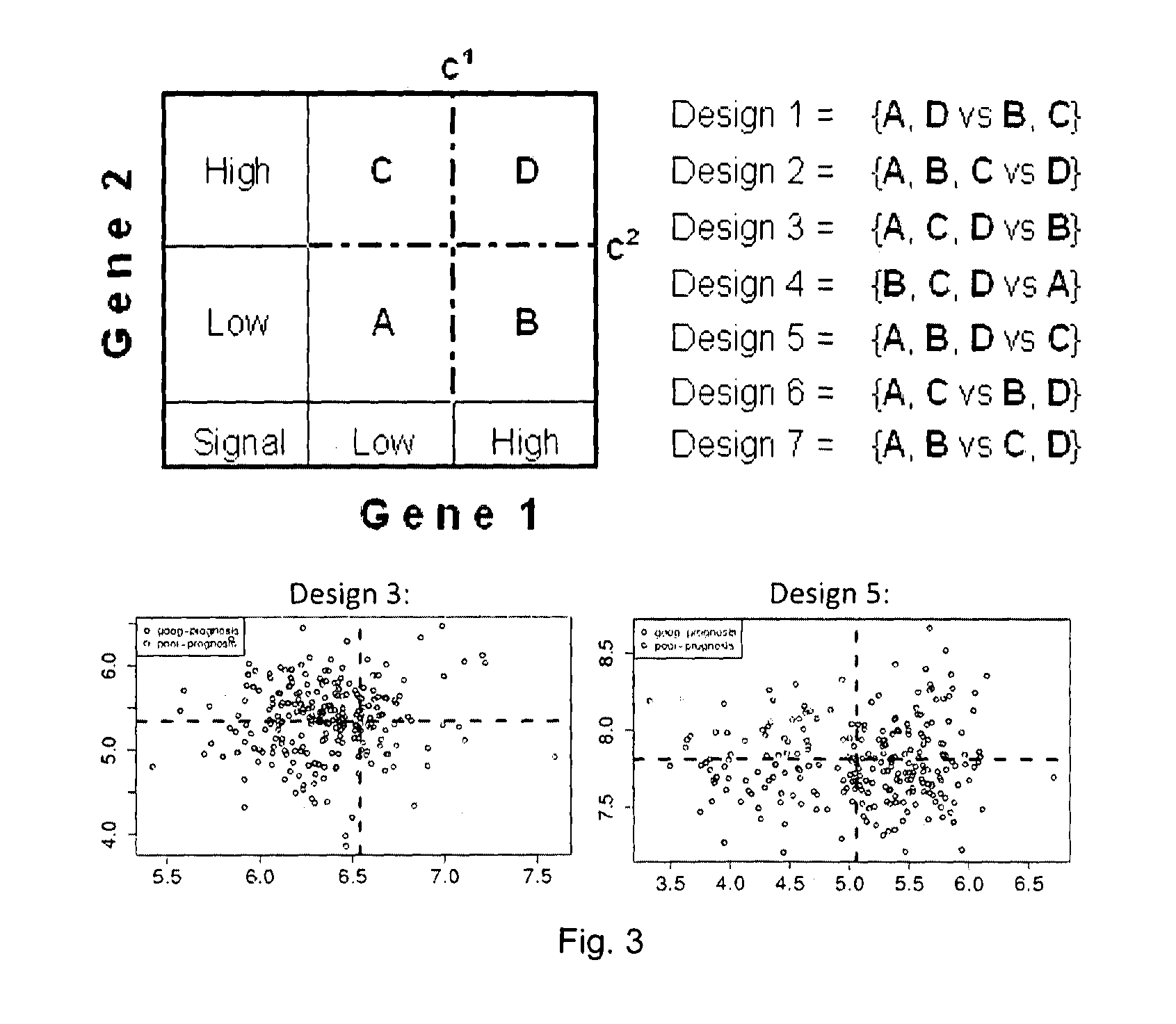
The present invention relates to a method of identification of clinically and genetically distinct sub-groups of patients subject to a medical condition, particularly breast, lung, and colon cancer patients using a composition of respective gene expression values for certain gene pairs. Sense-antisense gene pairs (SAGPs) which are relevant for a medical condition and the disease prognosis are used by the method to generate statistical models based on the expression values of the SAGPs. SAGPs for which the statistical models are found to have high value in prognosis of the variation of medical condition and the diseases are selected and integrated in the prognostic signature including specified parameters (e.g. cut-off values) of the prognostic model. It further relates to using respective gene expression values for these genes to predict patient′ risk groups (in context of patient's survival or / and disease progression) and to using the predicted groups for identification of patient risk, and specific and robust prognostic biomarkers with mechanistic interpretations of biological changes (associated with the gene signatures) appropriating for an implementation of therapeutic targeting.
Owner:AGENCY FOR SCI TECH & RES
Prognostic Marker for Endometrial Carcinoma
InactiveUS20110217701A1Microbiological testing/measurementDisease diagnosisNon small lung cancerRegimen
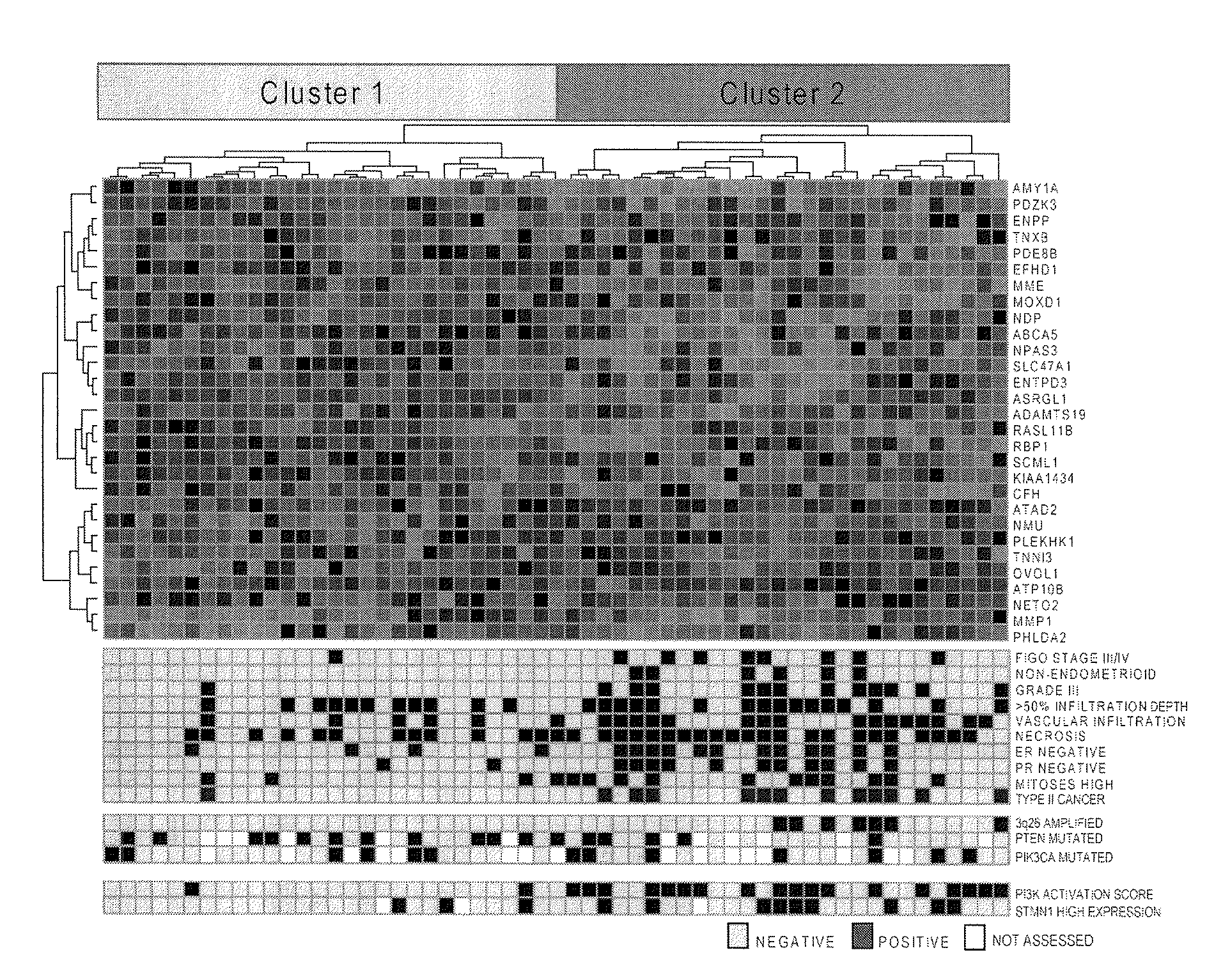
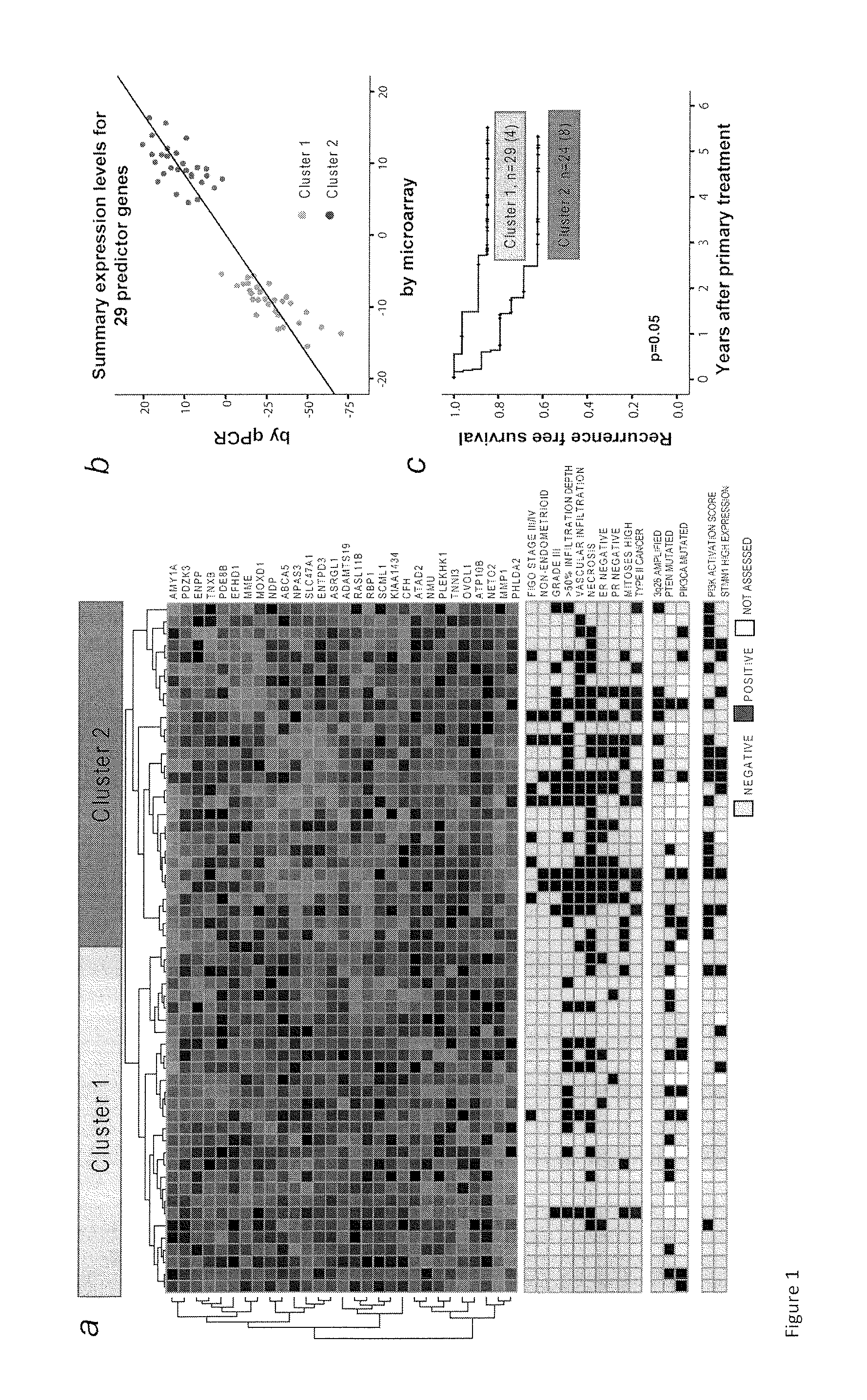
The present invention relates to a method for diagnosis of different stages of endometrial cancer in an individual. Further, the present invention relates to a method for evaluating the probability of survival for an individual suffering from endometrial carcinoma. In another aspect, the present invention relates to the stratification of therapy regimen of endometrial tumor, ovarian cancer, breast cancer, non-small lung cancer or hormone refractory prostate cancer therapy in an individual or monitoring therapeutic efficacy in an individual suffering from the same based on the expression status of STMN1 gene or protein. Moreover, the present invention relates to a kit for use in any of the above referenced methods comprising a means for determining amplifications and deletions of chromosomal regions 3q26.32 and 12p12.1, determining alterations of the gene expression profile of the genes (gene signature): upregulation of the genes PLEKHK1, ATP10B, NMU, MMP1, ATAD2, NETO2, TNNI3, PHLDA2, OVOL1 and down-regulation of the genes: NDP, KIAA1434, MME, CFH, MOXD1, SLC47A1, RBP1, PDE8B, ASRGL1, ADAMTS19, EFHD1, ABCA5, NPAS3, SCML1, TNXB, ENTPD3, AMY1A, ENPP, RASL11B, PDZK3, or the expression status of the STMN1 gene or protein, respectively. Finally, the present invention provides a method for predicting the response to taxanes in an individual suffering from a disease treated with the taxanes based on the expression status of the STMN1 gene or protein.
Owner:BERGEN TEKNOLOGIOVERFORING
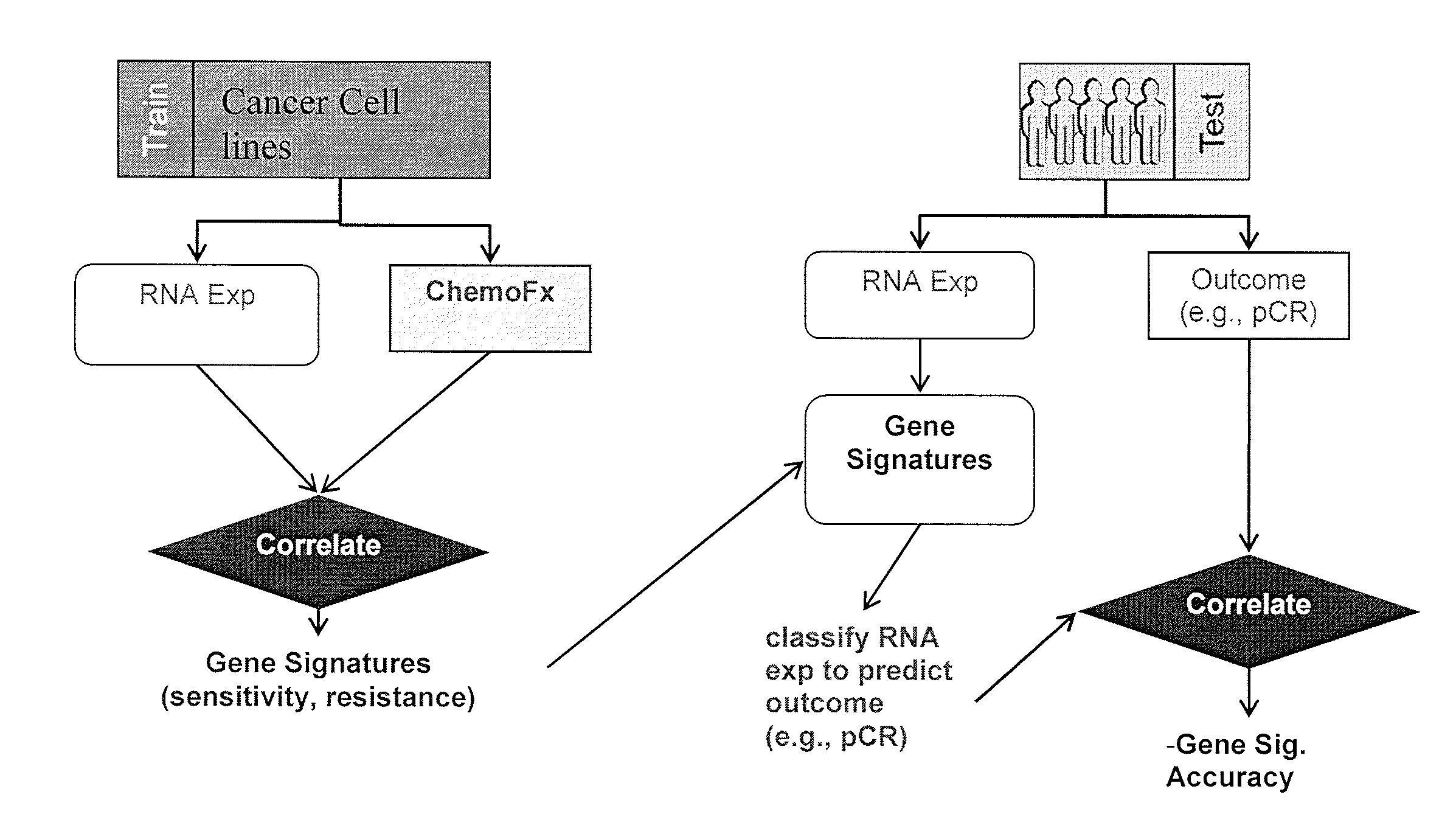
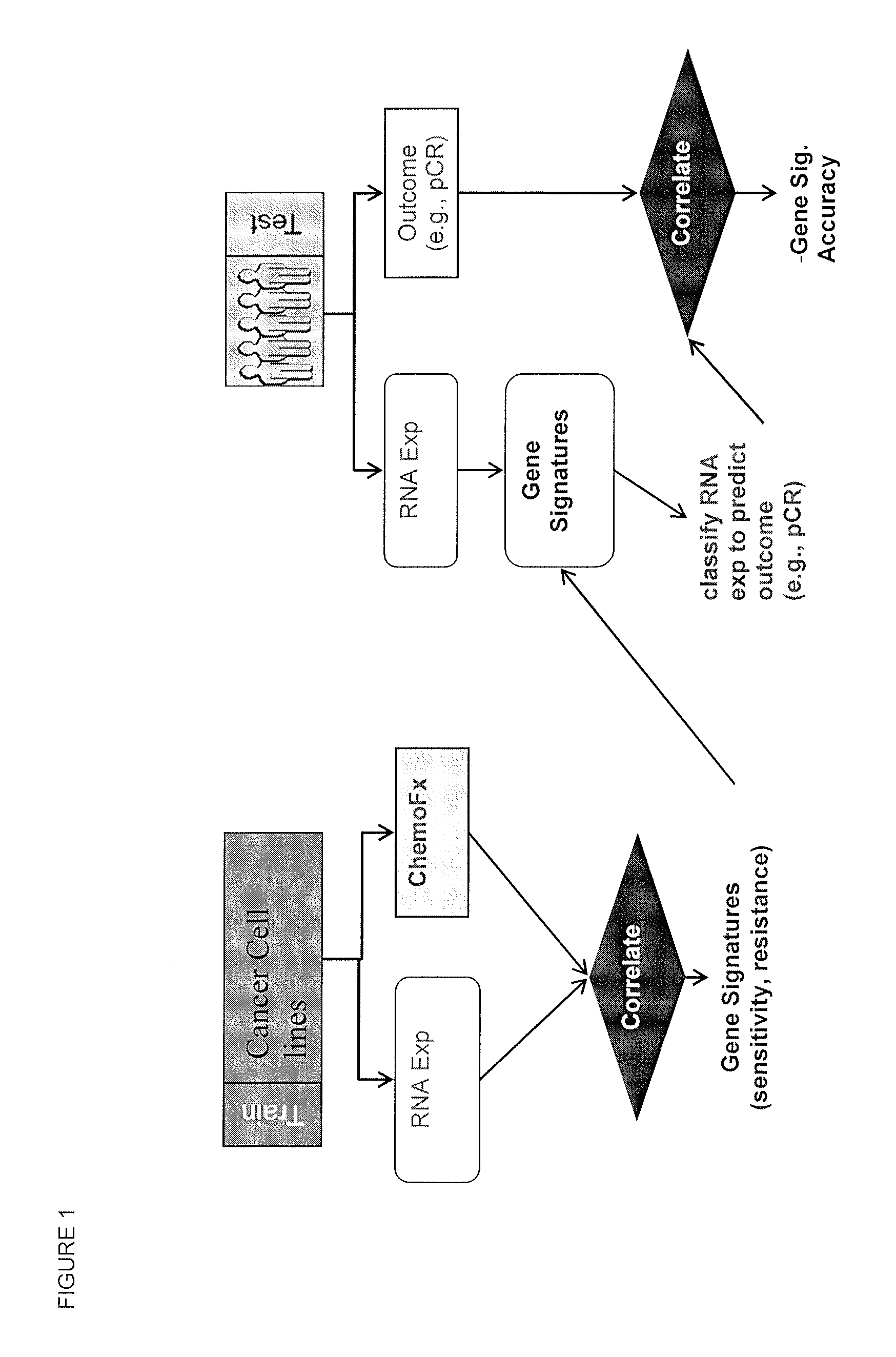
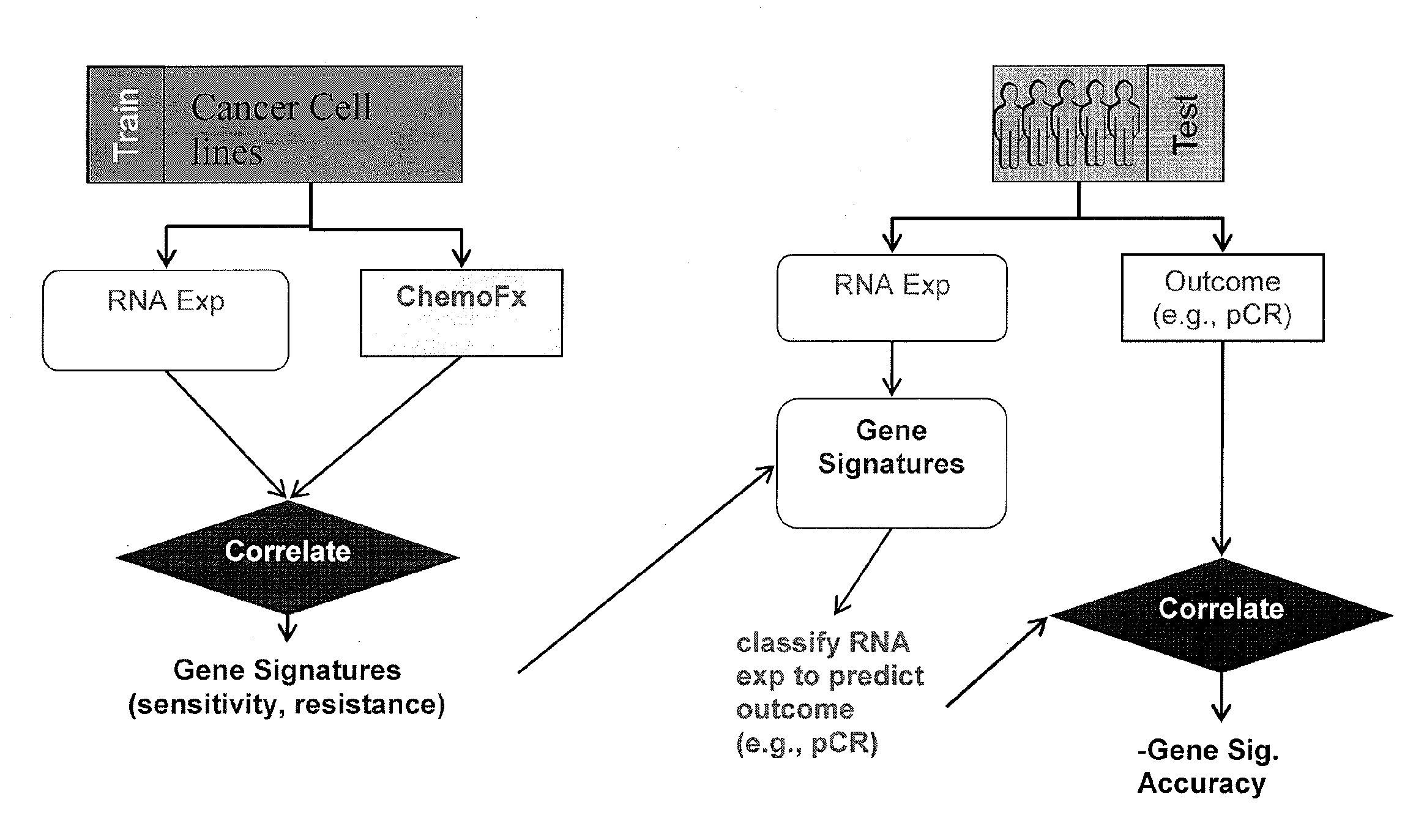
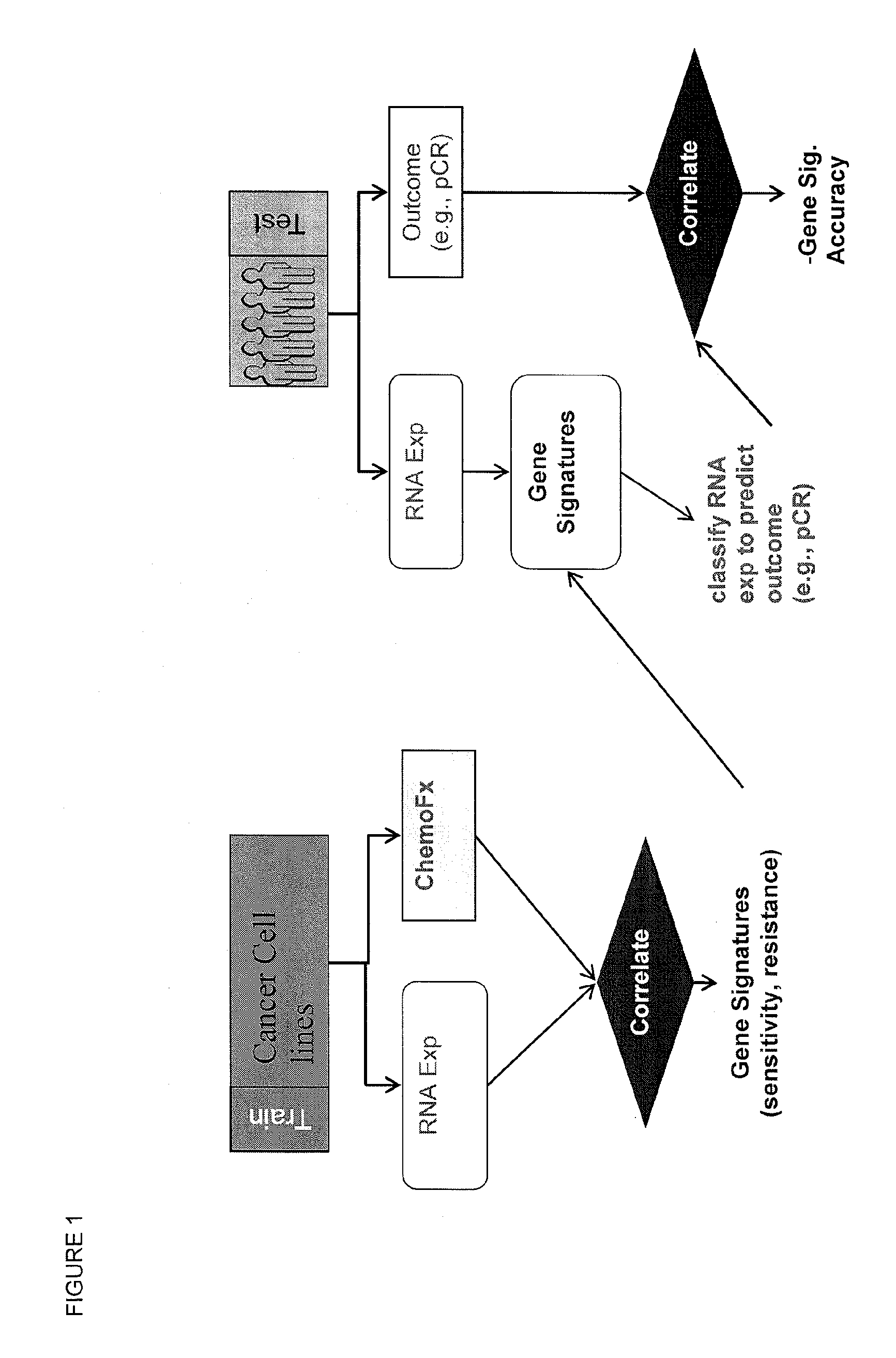
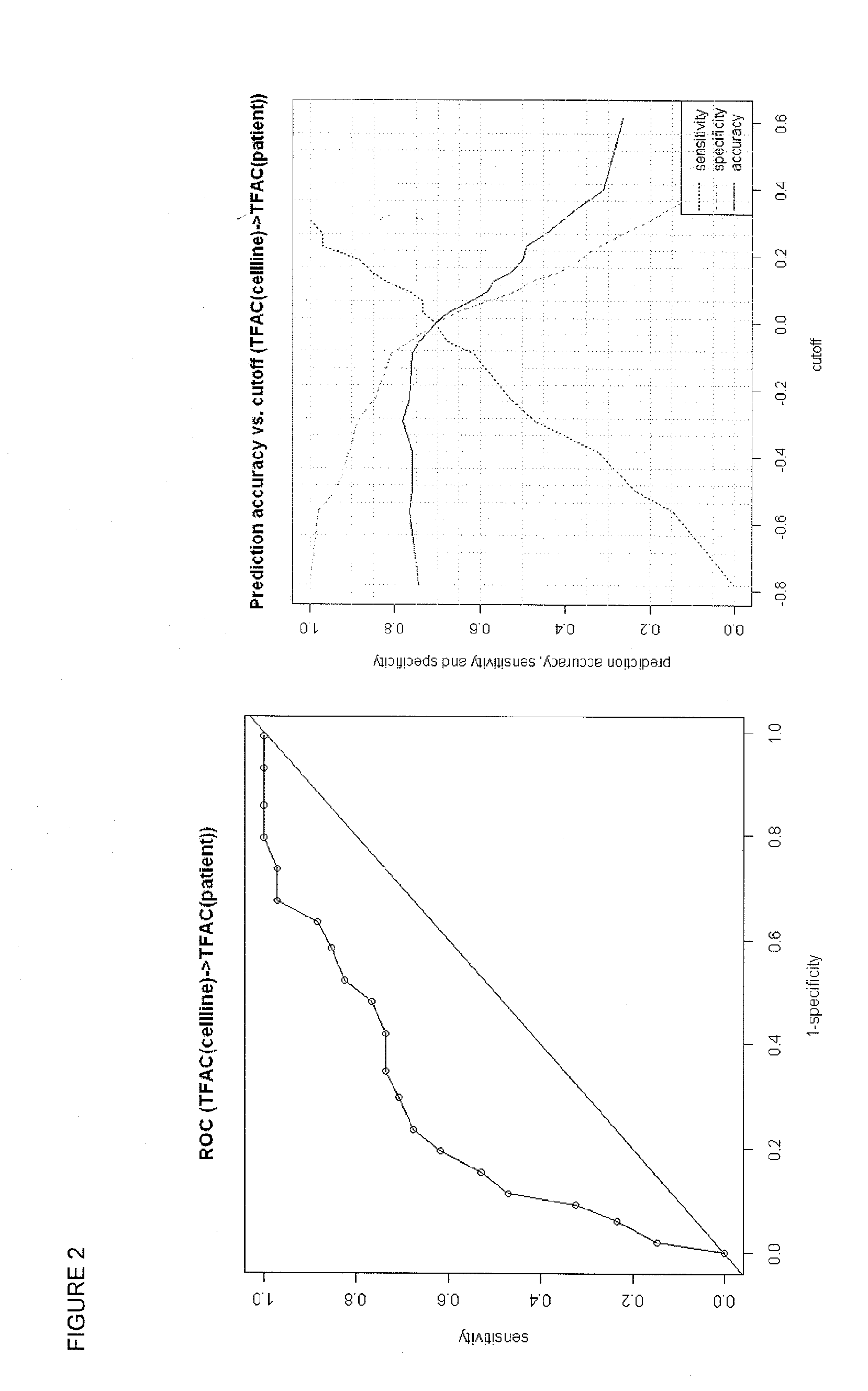
Methods and systems for evaluating the sensitivity or resistance of tumor specimens to chemotherapeutic agents
InactiveUS20120214679A1Complete and/or accurate prognostic and/or predictiveShorten the lengthNucleotide librariesMicrobiological testing/measurementAbnormal tissue growthRegimen
The present invention provides methods, systems, and kits for evaluating the sensitivity and / or resistance of tumor specimens to one or a combination of chemotherapeutic agents. Particularly, the invention provides malignant cell gene signatures that are predictive of a tumor's response to candidate chemotherapeutic regimens.
Owner:PRECISION THERAPEUTICS
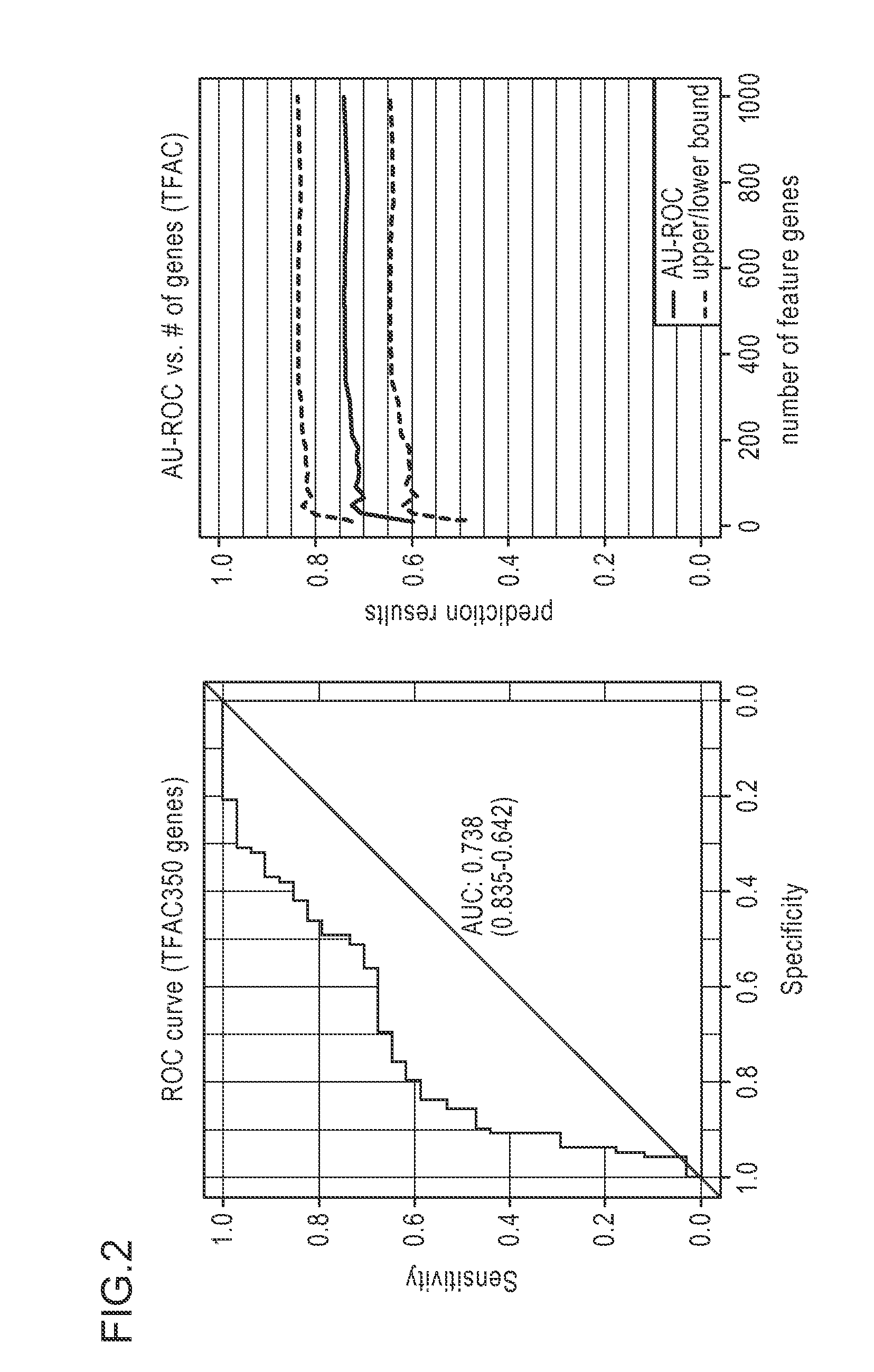
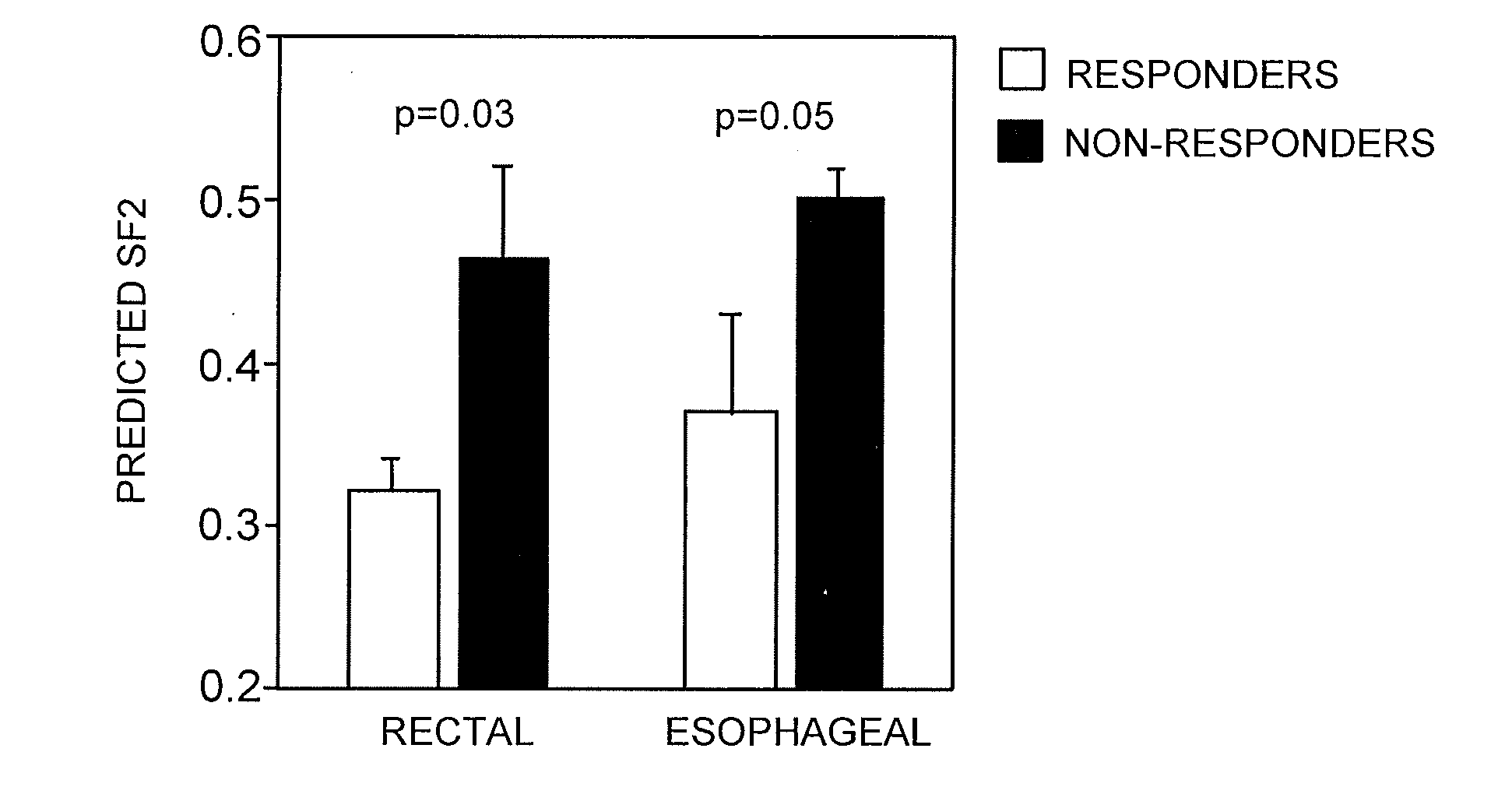
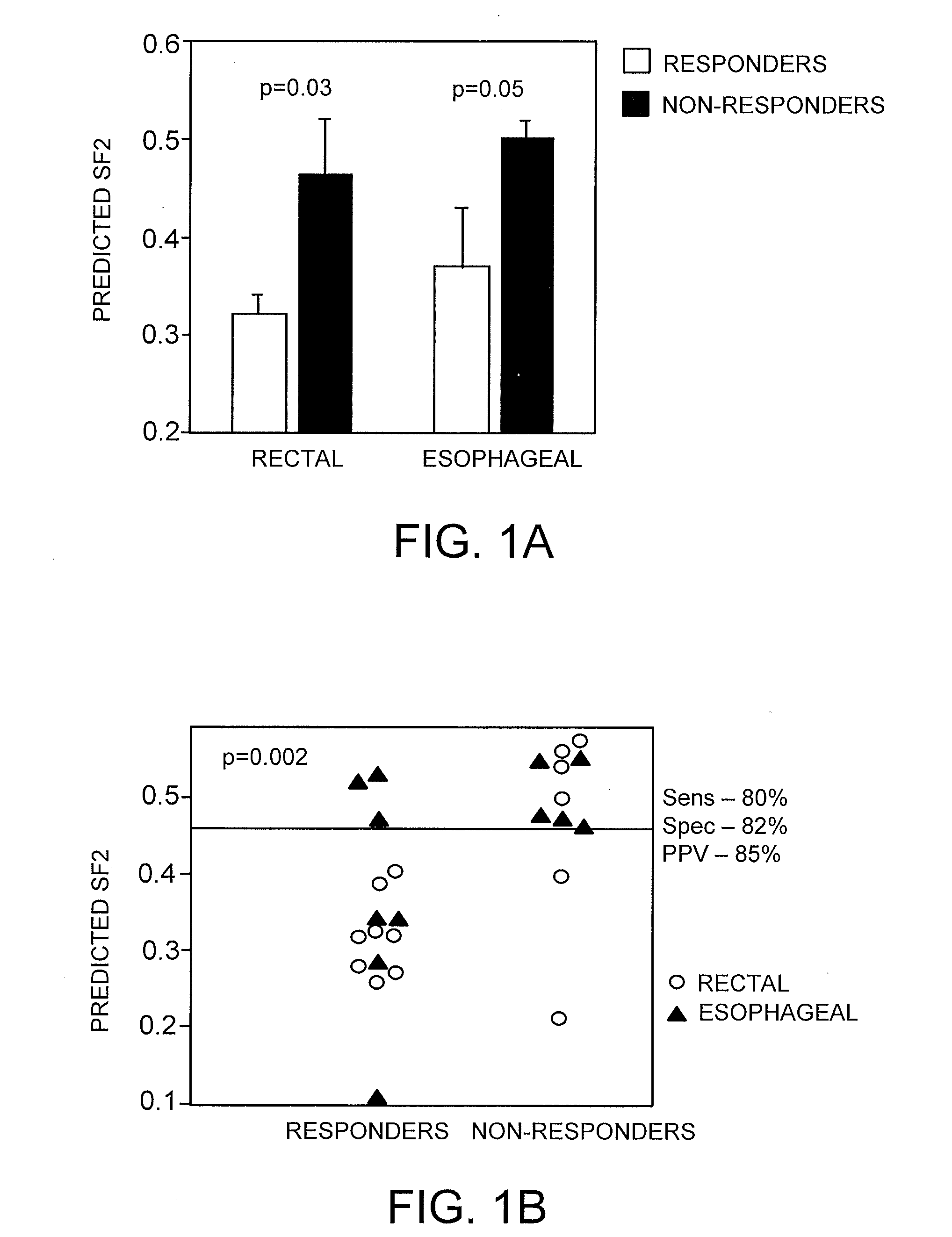
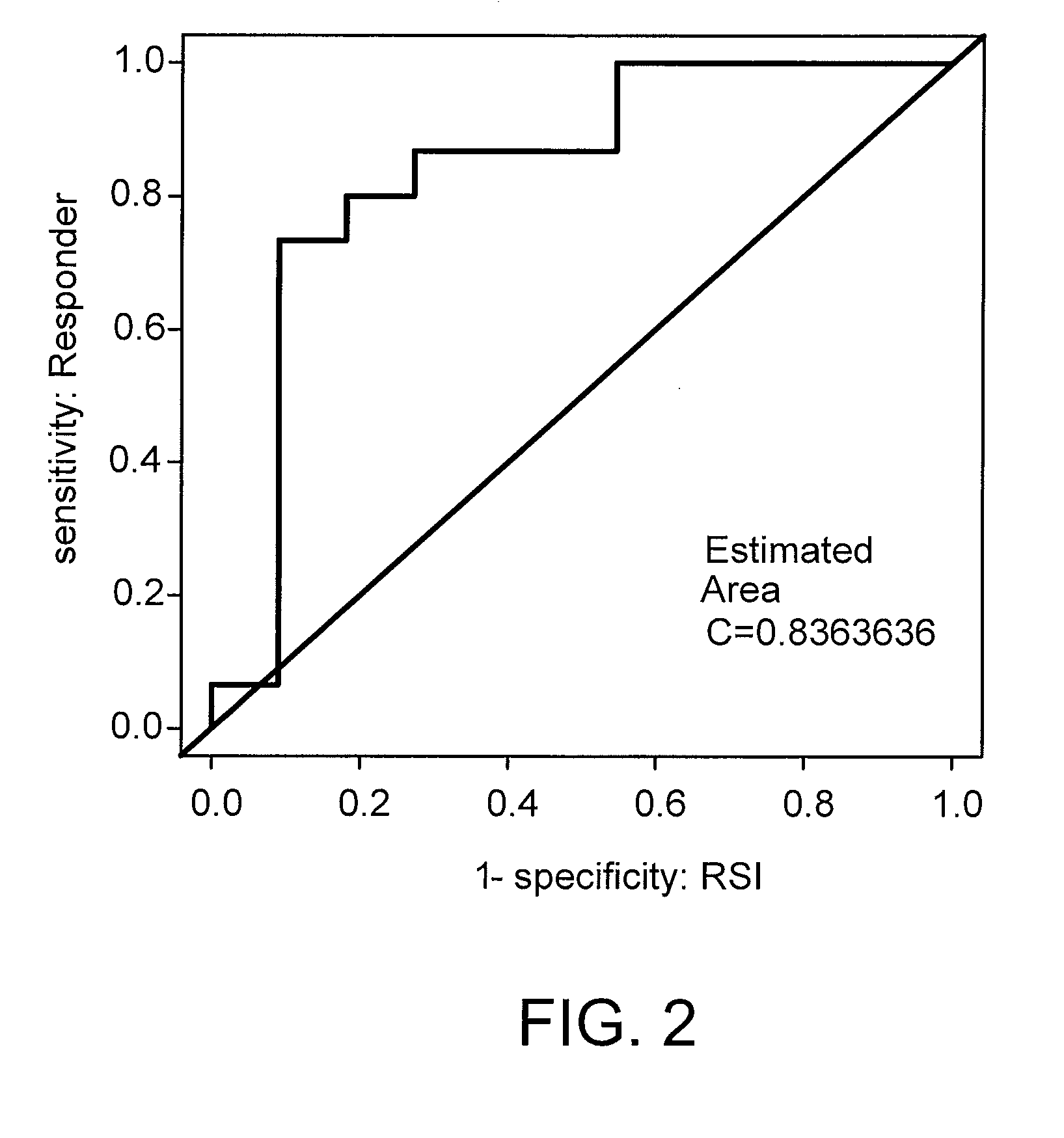
Gene Signature for the Prediction of Radiation Therapy Response
InactiveUS20090076734A1Increase and decrease in radiosensitivityIncrease chanceMedical simulationMicrobiological testing/measurementMathematical modelRadiation therapy
Described are mathematical models and method, e.g., computer-implemented methods, for predicting tumor sensitivity to radiation therapy, which can be used, e.g., for selecting a treatment for a subject who has a tumor.
Owner:UNIV OF SOUTH FLORIDA
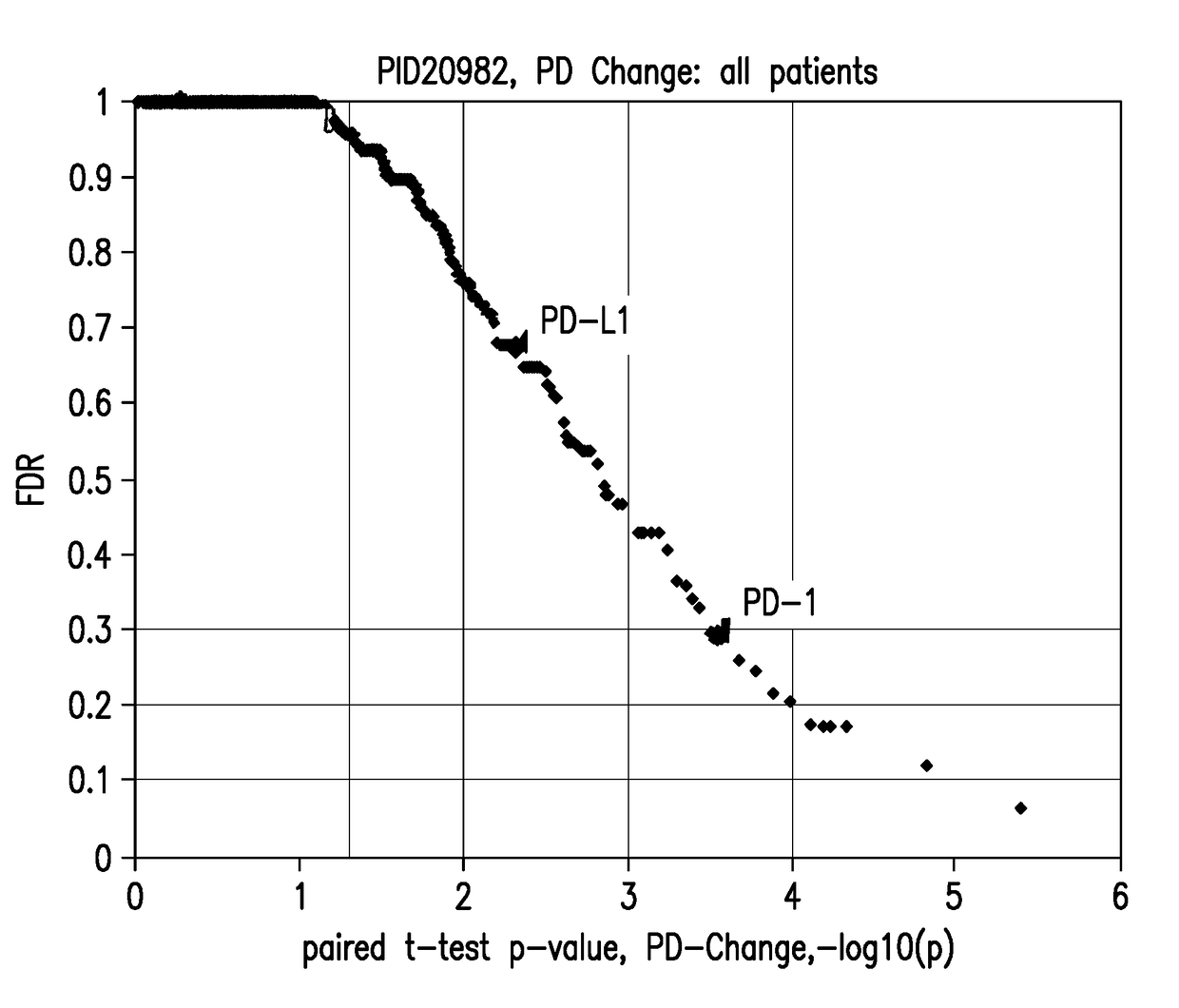
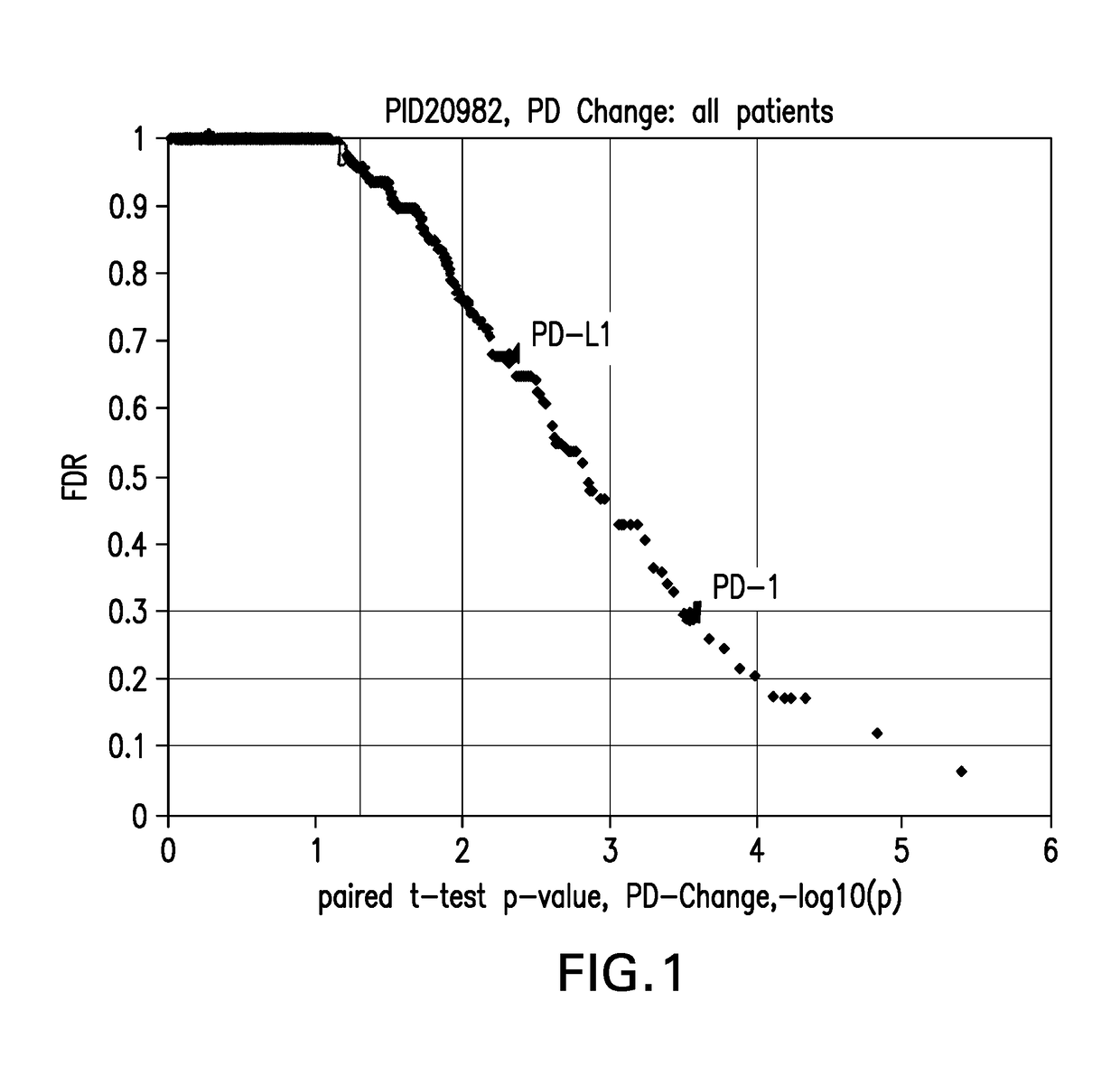
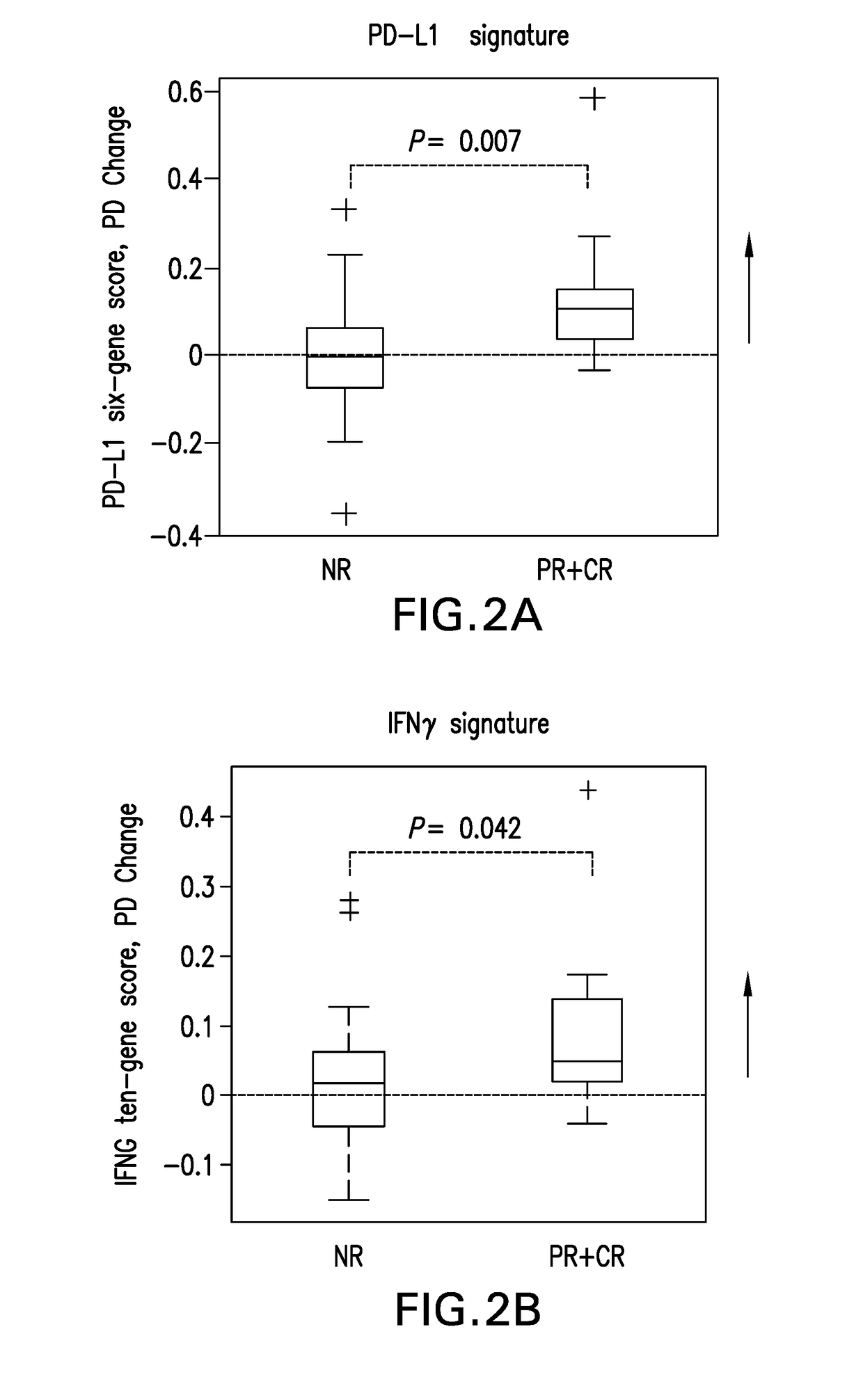
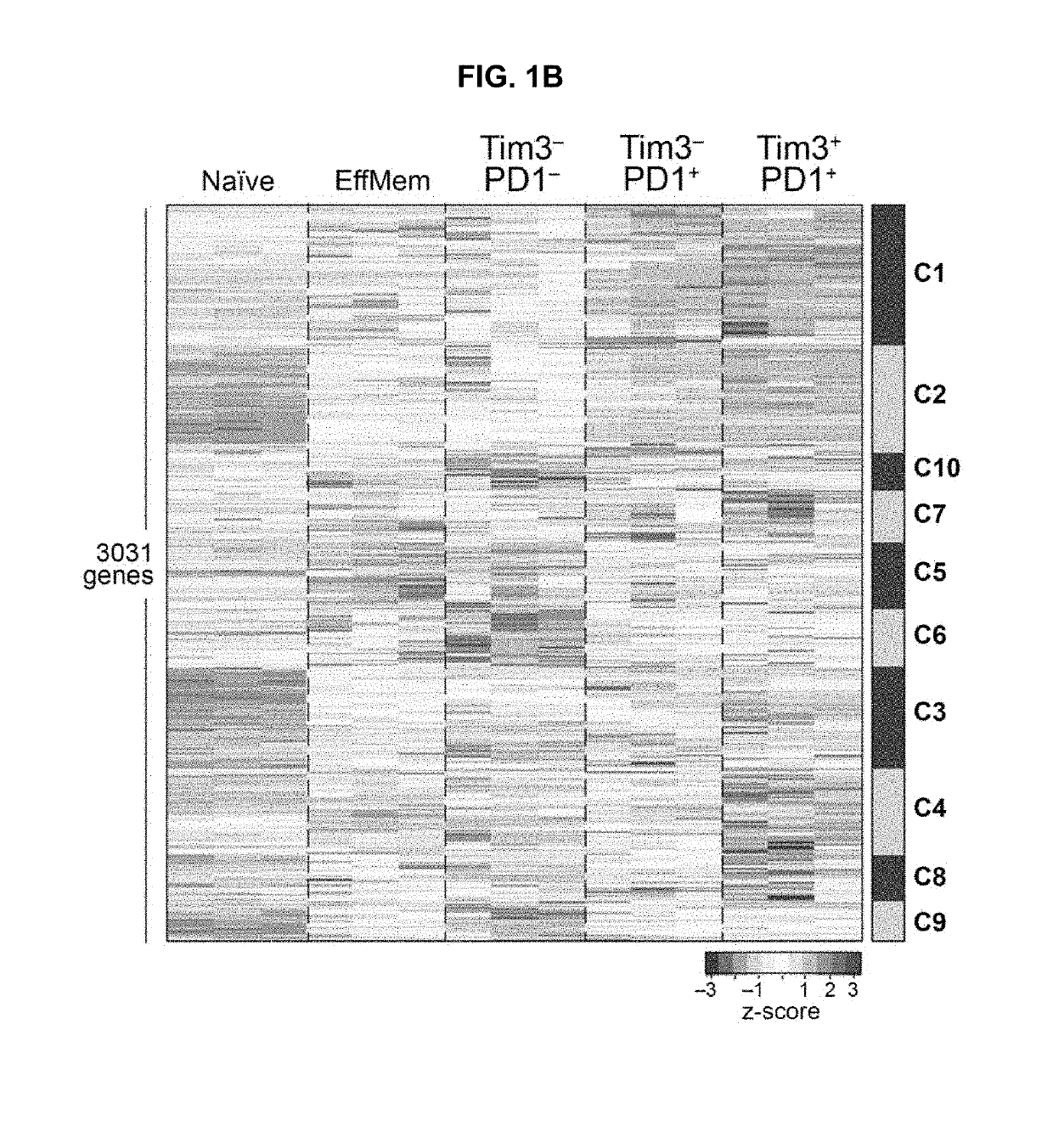
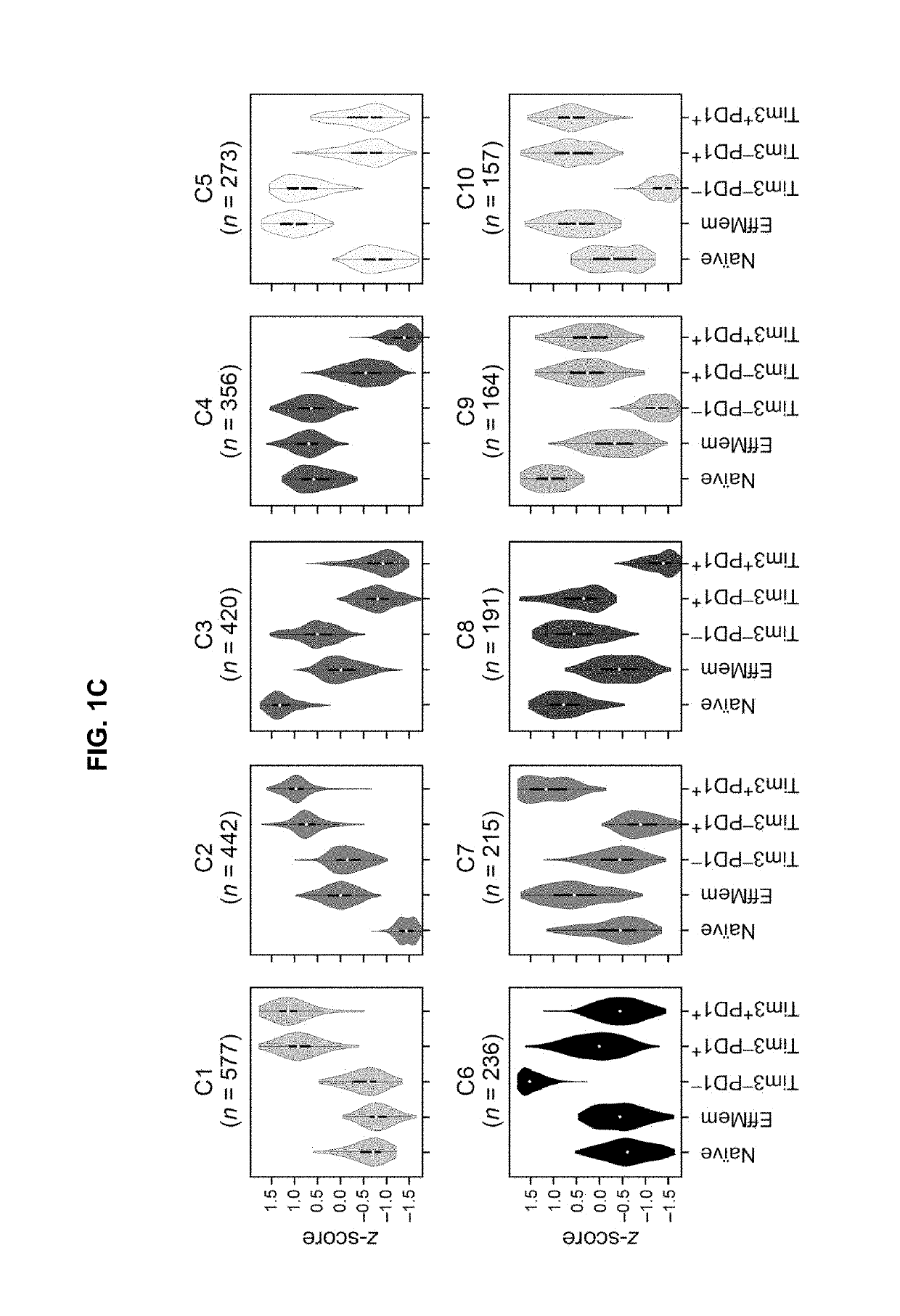
Blood-based biomarkers of tumor sensitivity to pd-1 antagonists
ActiveUS20180148790A1Inhibit bindingMicrobiological testing/measurementDisease diagnosisPhosphorylationPD-L1
The present disclosure describes baseline and on treatment blood-based gene signature biomarkers that are predictive of tumor sensitivity to therapy with a PD-1 antagonist. The on-treatment biomarkers comprise a PD-L1 gene signature or an interferon gamma gene signature and the baseline gene signature biomarker comprises genes associated with the oxidative phosphorylation pathway. The disclosure also provides methods and kits for testing tumor samples for these biomarkers, as well as methods for treating subjects with a PD-1 antagonist based on the test results.
Owner:MERCK SHARP & DOHME LLC
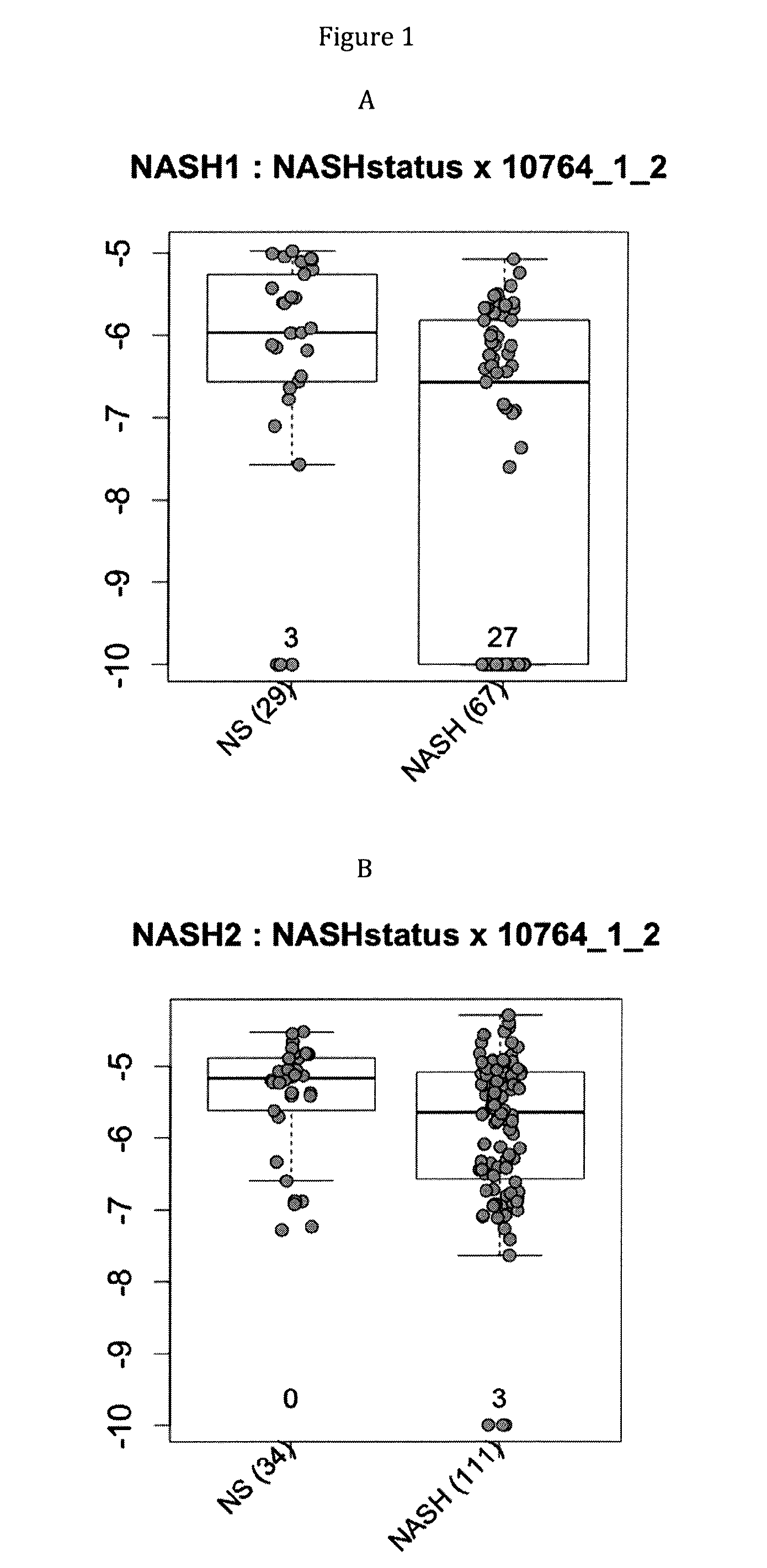
Gene signatures of inflammatory disorders that relate to the liver
ActiveUS20160068890A1Microbiological testing/measurementLibrary screeningIntestinal microorganismsUlcerative colitis
This invention is related to the area of characterization of inflammation in relation with the gut microbiota, in metabolic and autoimmune disorders. In particular, it relates to the identification of gene signatures which can be used as a marker predictive of inflammation associated diseases, such as liver-related metabolic disorders, in particular to the evolution of benign steatosis towards its most severe forms (steatohepatitis and cirrhosis) or autoimmune disorders, in particular inflammatory bowel diseases (Crohn's and Ulcerative Colitis). These gene signatures can therefore be used as a means of diagnosis, prognosis, stratification for drug studies, for monitoring patient and for assigning an appropriate treatment.
Owner:INST NAT DE RECH POUR LAGRICULTURE LALIMENTATION & LENVIRONNEMENT +1
Mucosal gene signatures
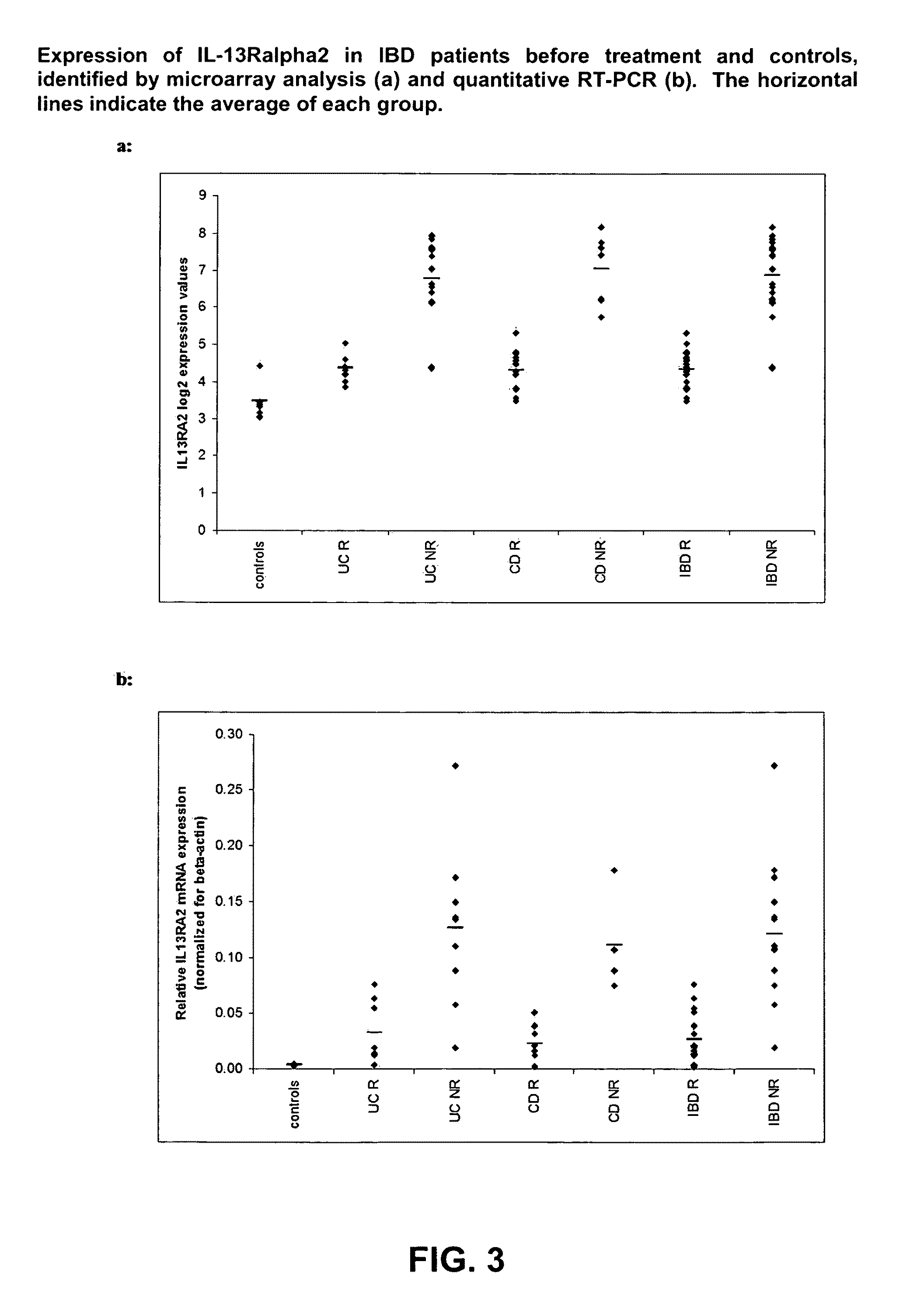
InactiveUS20110059445A1Inhibit expressionInhibitory activityMicrobiological testing/measurementImmunoglobulinsMucosal BiopsyUlcerative colitis
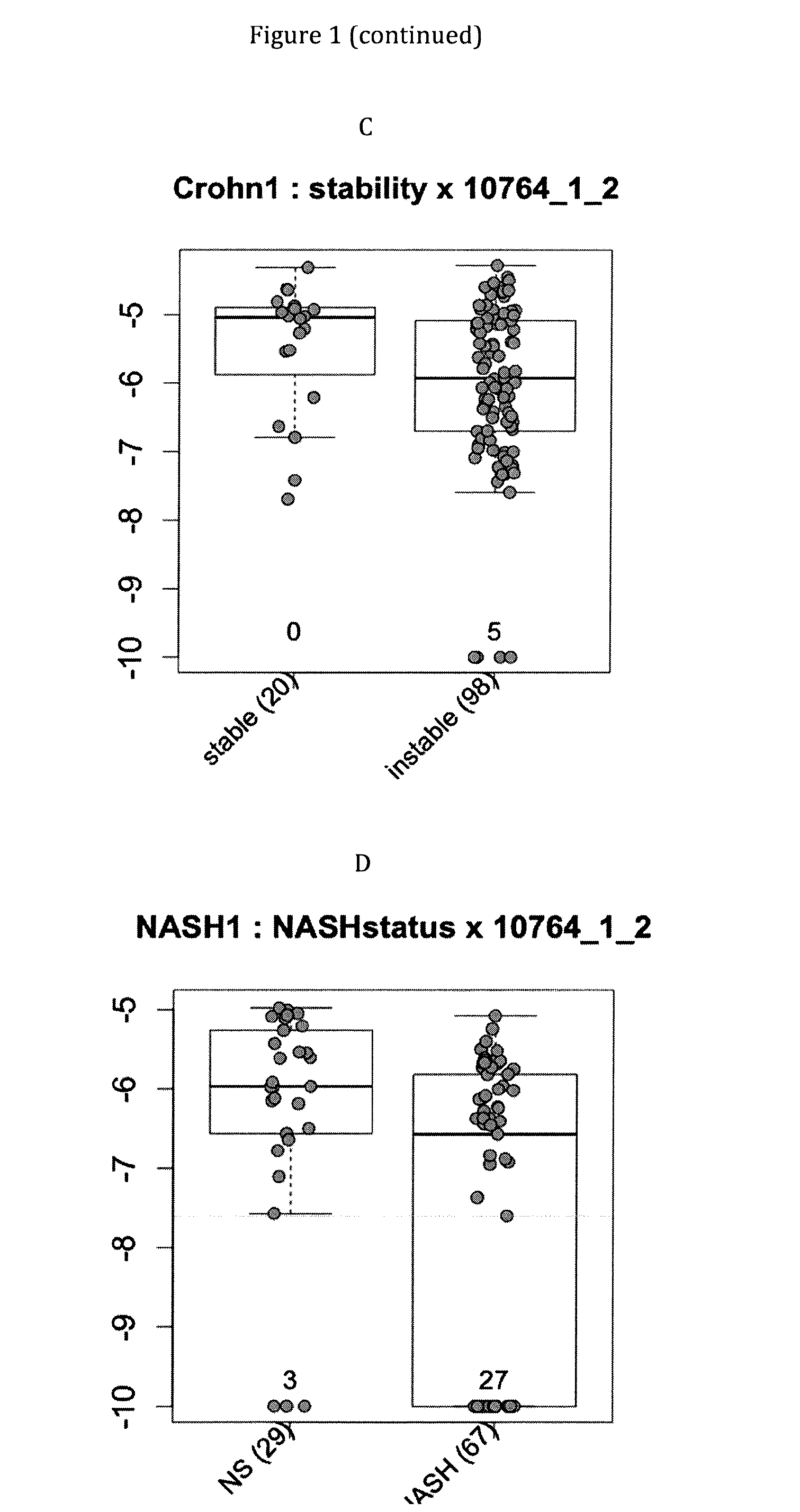
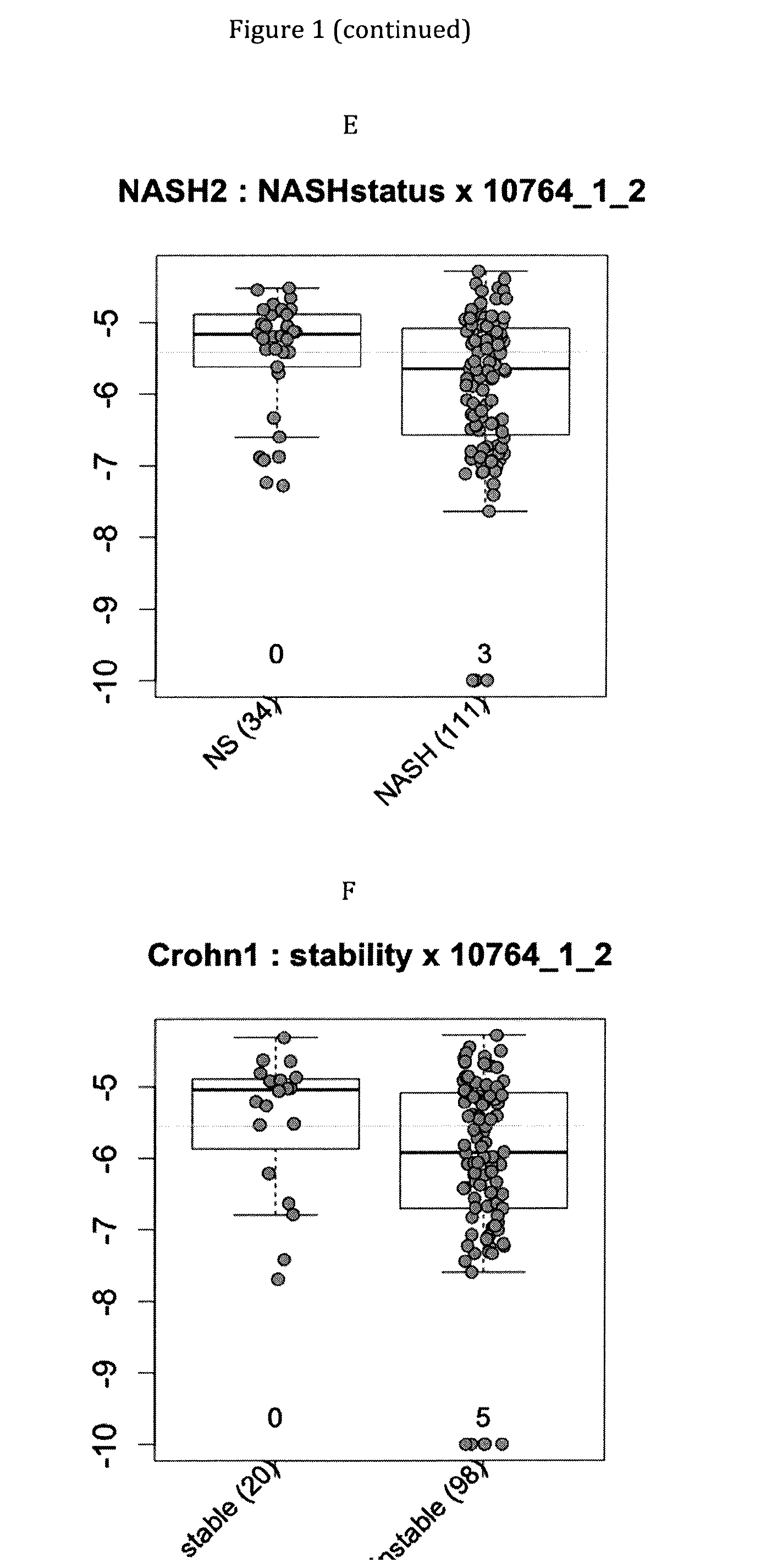
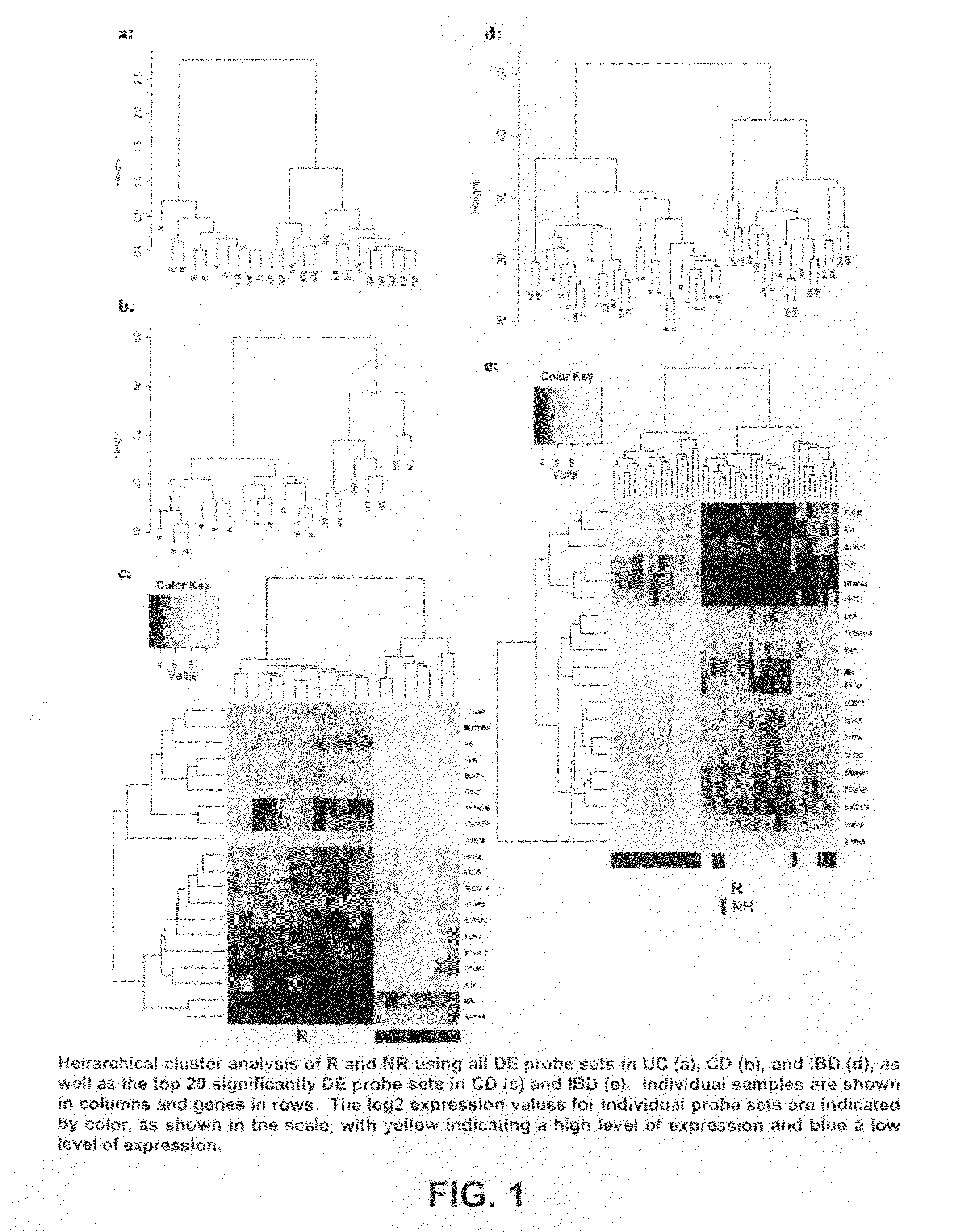
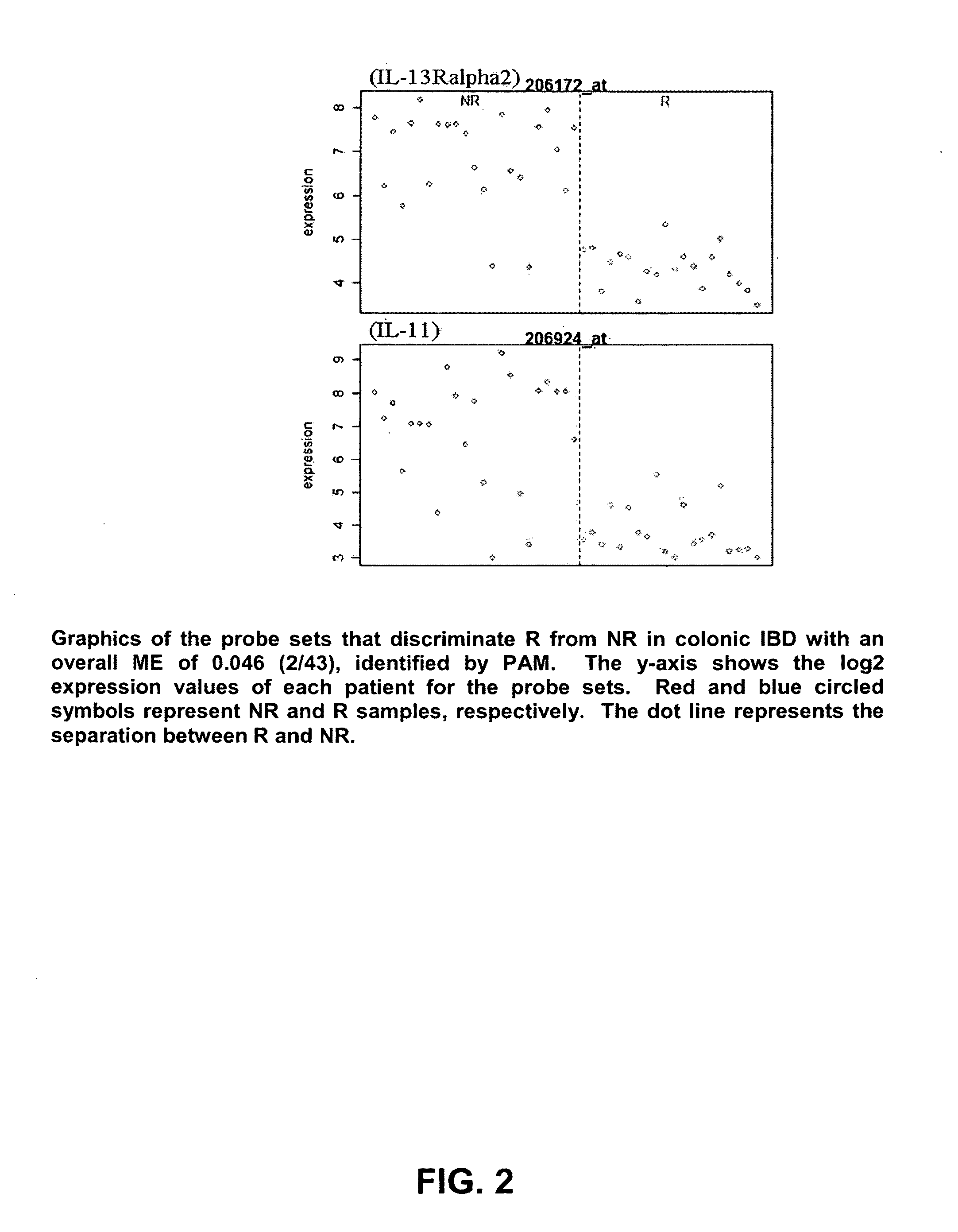
Infliximab (IFX) is an effective treatment for Crohn's disease (CD) and ulcerative colitis (UC) not responding to standard therapy. Thirty percent to forty percent of patients however do not improve and the response is often incomplete. We identified mucosal gene signatures predictive of response to EFX using high-density oligonucleotide arrays. Eight UC patients and twelve CD patients showed healing. In UC, only one probe set was differentially expressed in responders compared with non-responders, i.e., IL-13R(alpha)2. At PAM analysis, two probe sets, representing IL-13Ralpha2 and IL-I 1, separated IBD responders from non-responders with an overall misclassification error rate of 0.046 (2 / 43), with 100% sensitivity and 91.3% specificity. The IL-13R(alpha)2 probe set was a top-ranked probe set in all our analyses using both LIMMA and PAM strategies. Our gene array studies of mucosal biopsies identified IL-13R(alpha)2 in IBD as a predictor of response or non-response to IFX.
Owner:KATHOLIEKE UNIV LEUVEN
Gene signature biomarkers of tumor response to pd-1 antagonists
InactiveUS20160312295A1Improve expression levelMicrobiological testing/measurementChemoinformaticsTumor responseTumor Sample
The present disclosure describes gene signature biomarkers that are useful for identifying cancer patients who are most likely to benefit from treatment with a PD-1 antagonist. The disclosure also provides methods and kits for testing tumor samples for the biomarkers, as well as methods for treating subjects with a PD-1 antagonist based on the test results.
Owner:MERCK SHARP & DOHME CORP
Information management systems and methods using a biological signature
InactiveCN104769134AEliminate fragmentationEasy to integrateMicrobiological testing/measurementBiostatisticsDigital signatureComputer security
Systems and methods are provided for generating a biological signature such as a genetic signature and using such signature of an individual. The biological signature may be used to verify the identity of the individual. A verified individual may be granted access to a secured location, item, and / or service. Biological signatures may also be used to search or aggregate records for an individual.
Owner:THERANOS IP CO LLC
Methods and systems for evaluating the sensitivity or resistance of tumor specimens to chemotherapeutic agents
InactiveUS20100331210A1Precise processReduce in quantityNucleotide librariesMicrobiological testing/measurementRegimenWilms' tumor
The present invention provides methods, systems, and kits for evaluating the sensitivity and / or resistance of tumor specimens to one or a combination of chemotherapeutic agents. Particularly, the invention provides malignant cell gene signatures that are predictive of a tumor's response to candidate chemotherapeutic regimens.
Owner:PRECISION THERAPEUTICS
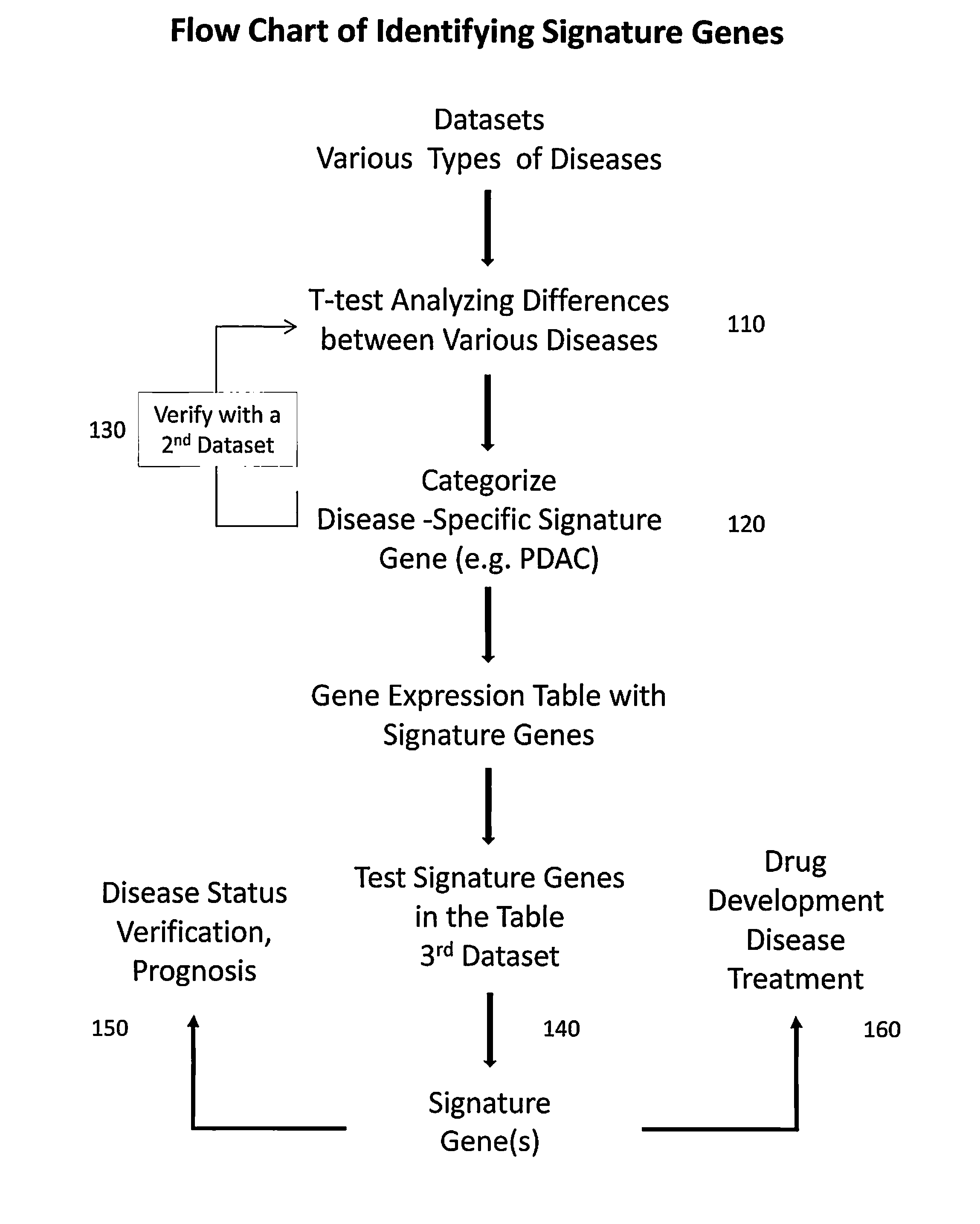
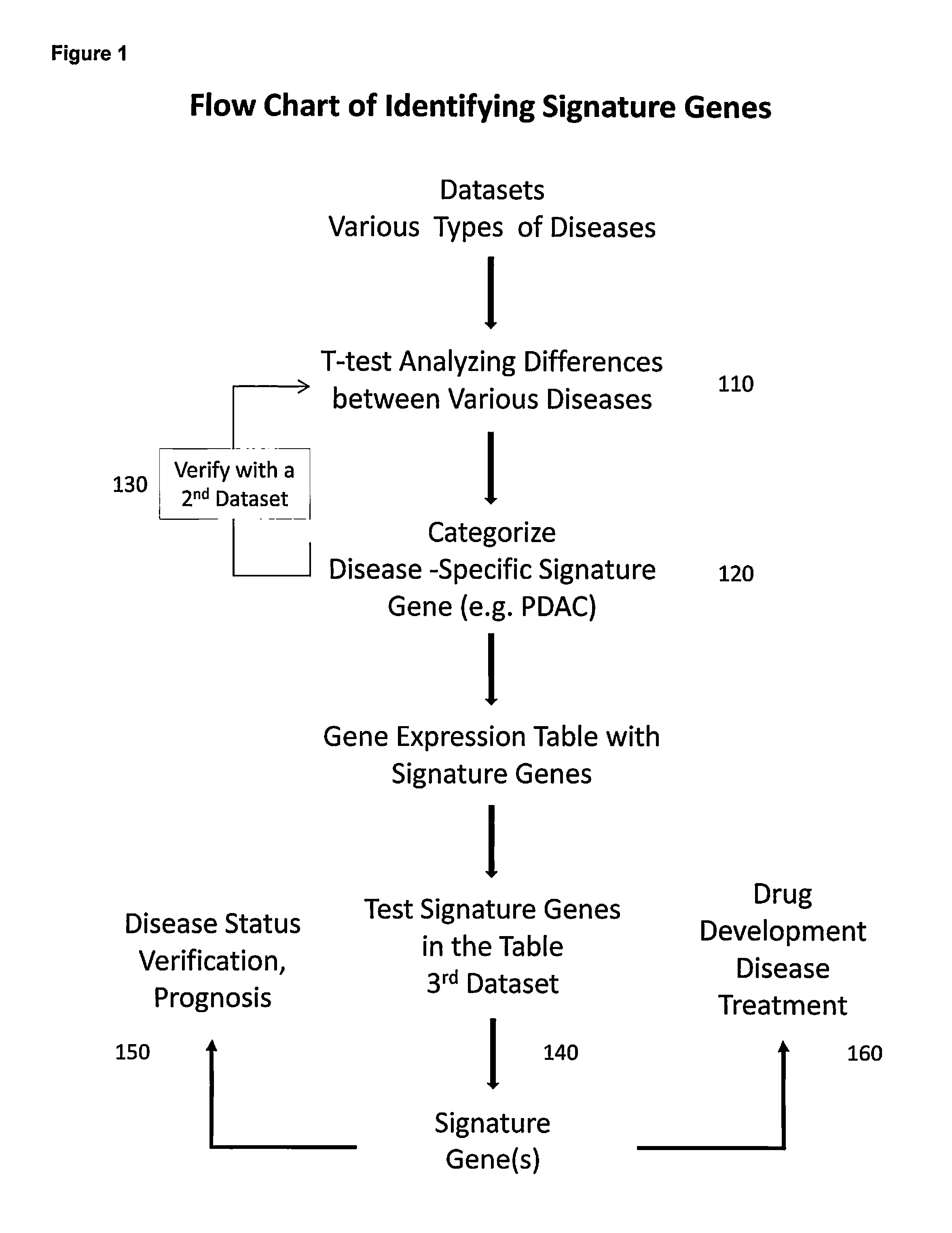
Gene signatures for detection of potential human diseases
A process to select signature gene by performing statistical analyses on gene datasets of various types of diseases for identifying signature genes for at least one of the diseases followed by categorizing and establishing gene expression table with the signature genes. The signature genes in the gene expression table are tested and verified by applying additional datasets to finalize and confirm the signature genes. The step of performing the statistical analyses on the gene datasets of various types of diseases further comprising a step of performing a total background normalization (TBN) of a relative gene expression (RGE) ratio then carried a two-tail T-test of the RGE ratios between the various diseases. The step of identifying the signature gene further comprising a step of carrying out a false positive elimination (FPE) by identifying differently expressed genes and removing overlapping genes among different diseases from a list of the signature genes
Owner:YANG YIH SHENG
Gene signatures associated with efficacy of postmastectomy radiotherapy in breast cancer
InactiveUS20150111758A1Improve expression levelHigh expressionPeptide librariesNucleotide librariesPostmastectomy radiotherapyCourse of action
The present invention relates to compositions, kits, and methods for providing a prognosis and / or determining a treatment course of action in a subject diagnosed with breast cancer. In particular, the present invention relates to gene expression signatures useful in the prognosis, diagnosis, and treatment of breast cancer.
Owner:UNIV OSLO HF +1
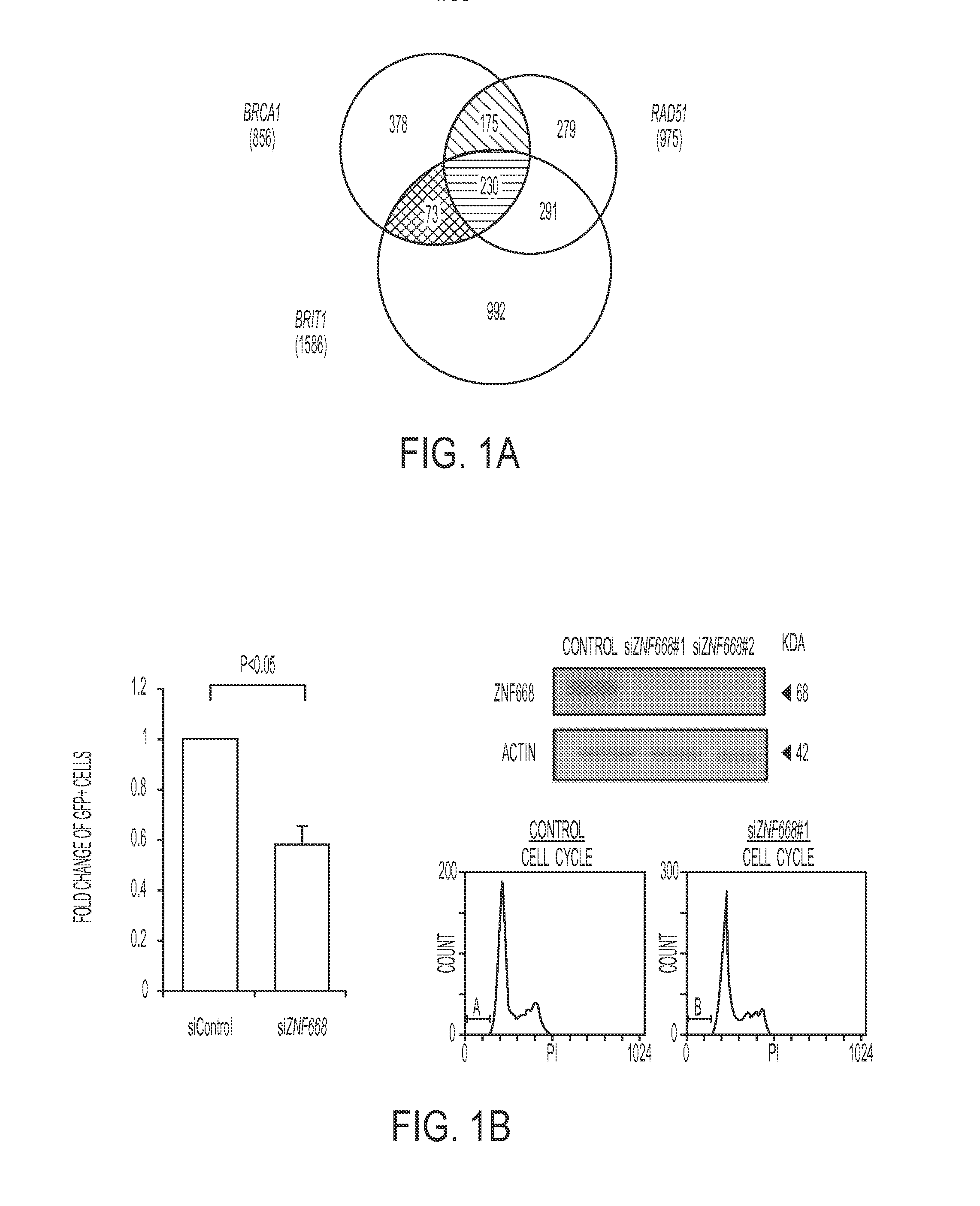
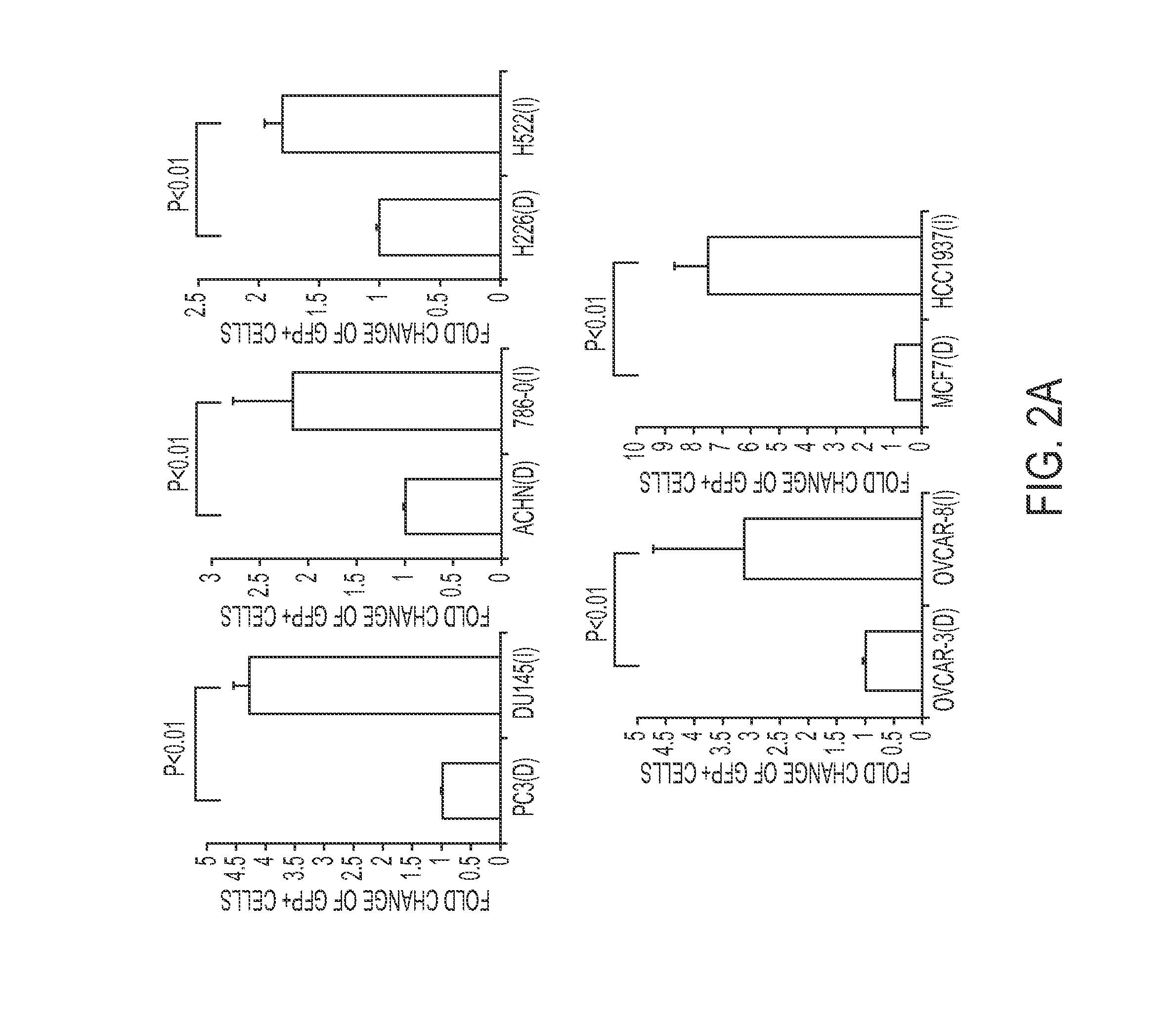
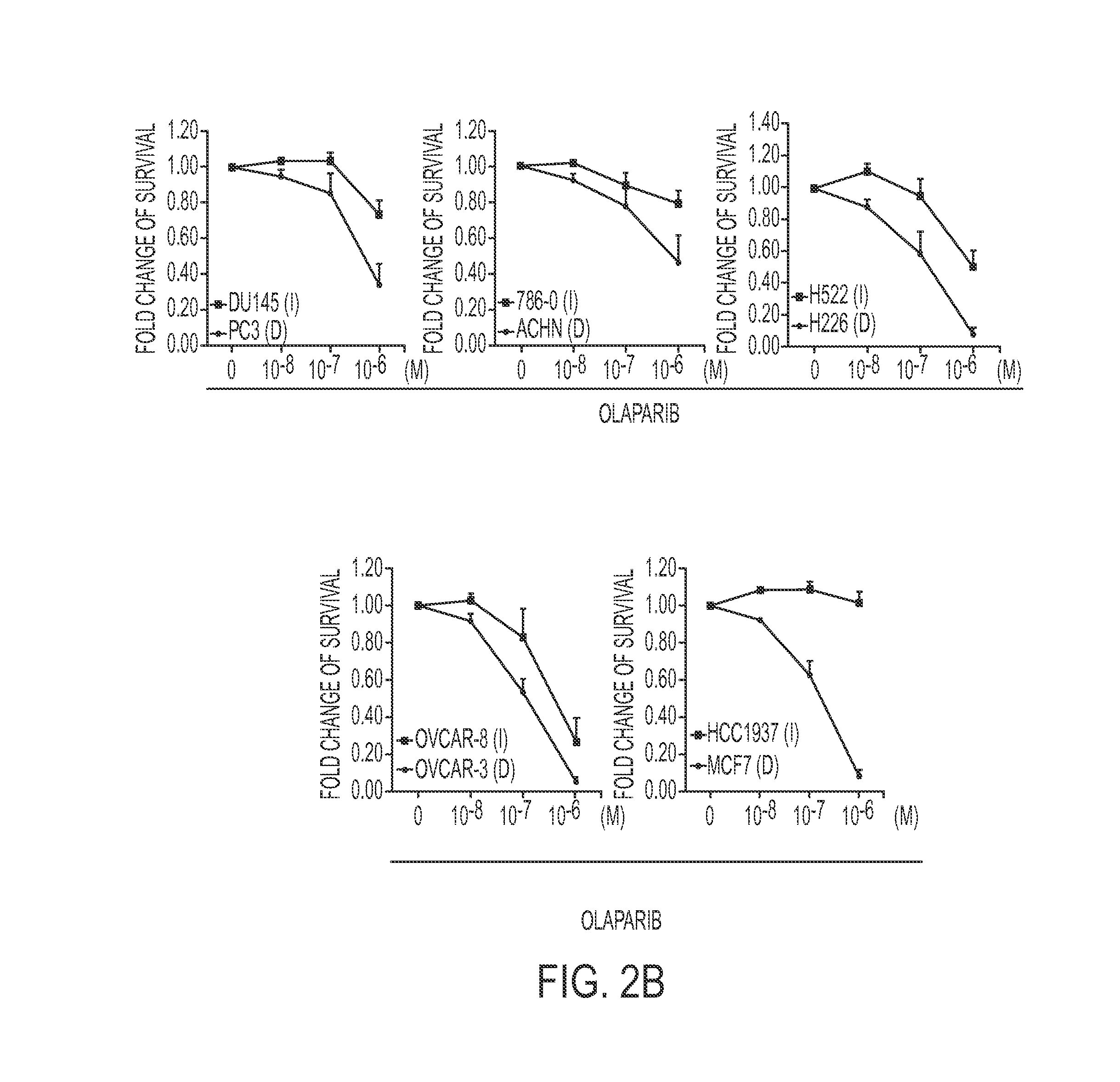
Gene signature to predict homologous recombination (HR) deficient cancer
Methods for identifying and treating cancers that are homologous recombination (HR)-repair defective. In some aspects, HR defective cancers are treated with a PARP inhibitor therapy. Methods for sensitizing cancers to a PARP inhibitor therapy are also provided.
Owner:BOARD OF RGT THE UNIV OF TEXAS SYST
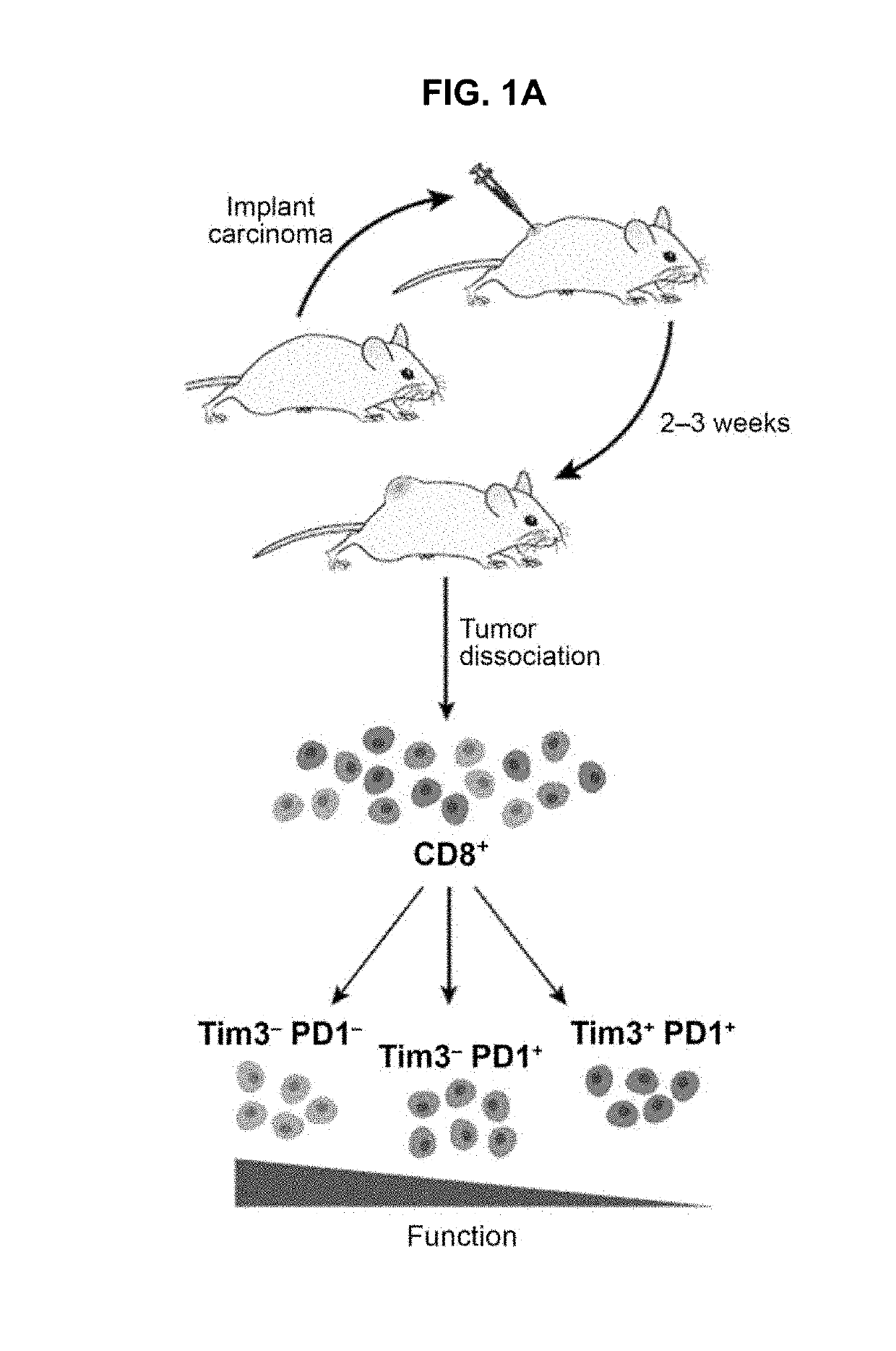
Compositions and methods for evaluating and modulating immune responses by use of immune cell gene signatures
PendingUS20190100801A1Lower immune responseEnhance immune responseMicrobiological testing/measurementImmune Cell FunctionGene
The present invention provides markers, marker signatures and molecular targets that correlate with dysfunction of immune cells and are advantageously independent of the immune cell activation status. The present markers, marker signatures and molecular targets provide for new ways to evaluate and modulate immune responses. Therapeutic methods are also provided to treat a patient in need thereof who would benefit from an increased immune response.
Owner:THE BROAD INST INC +2
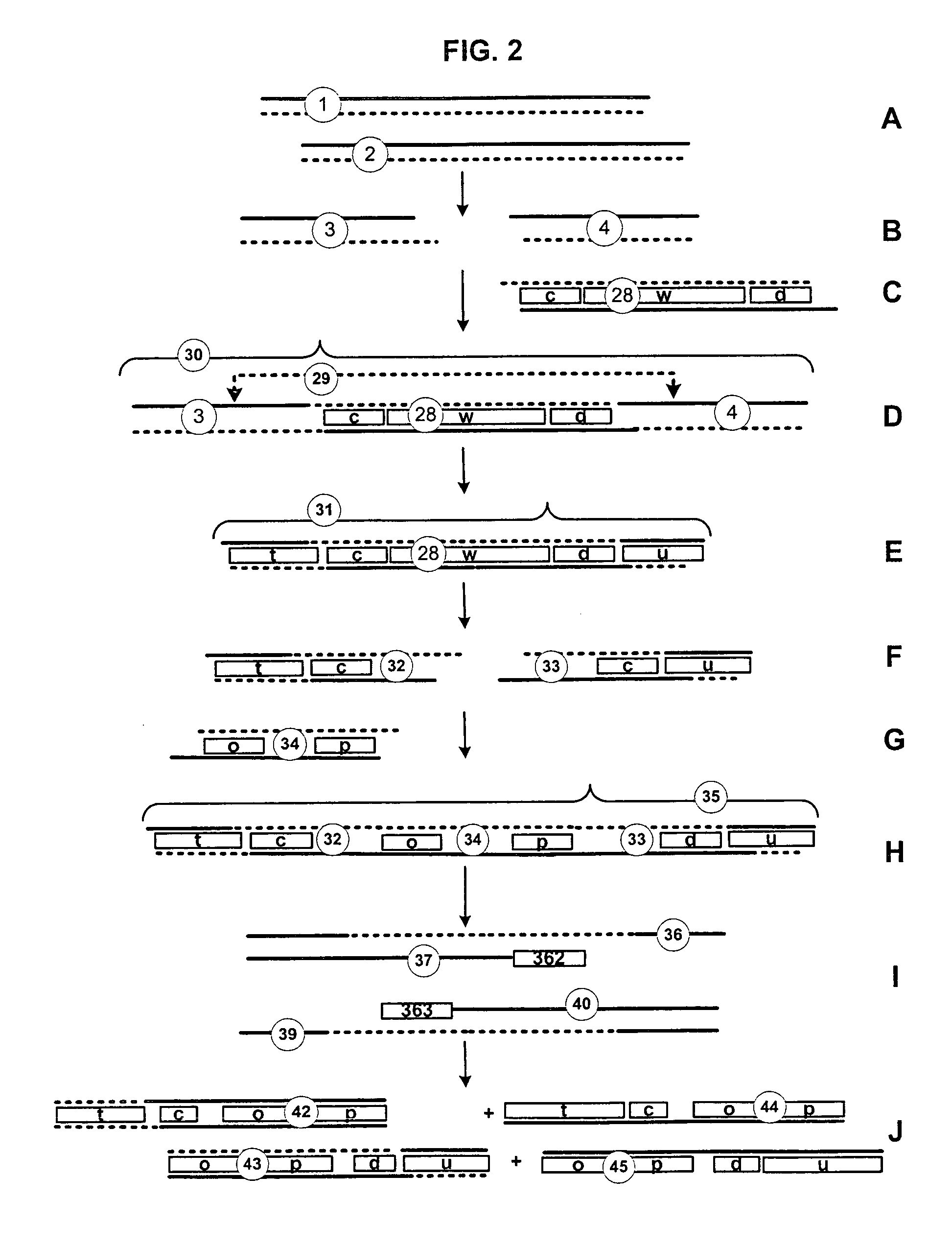
Producing, cataloging and classifying sequence tags
InactiveUS7618778B2Overcome limitationsShortening the linear chimeric nucleic acid intermediatesSugar derivativesMicrobiological testing/measurementCatalogingDouble strand
Owner:KAUFMAN JOSEPH C
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com