Methods for predicting cancer outcome and gene signatures for use therein
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
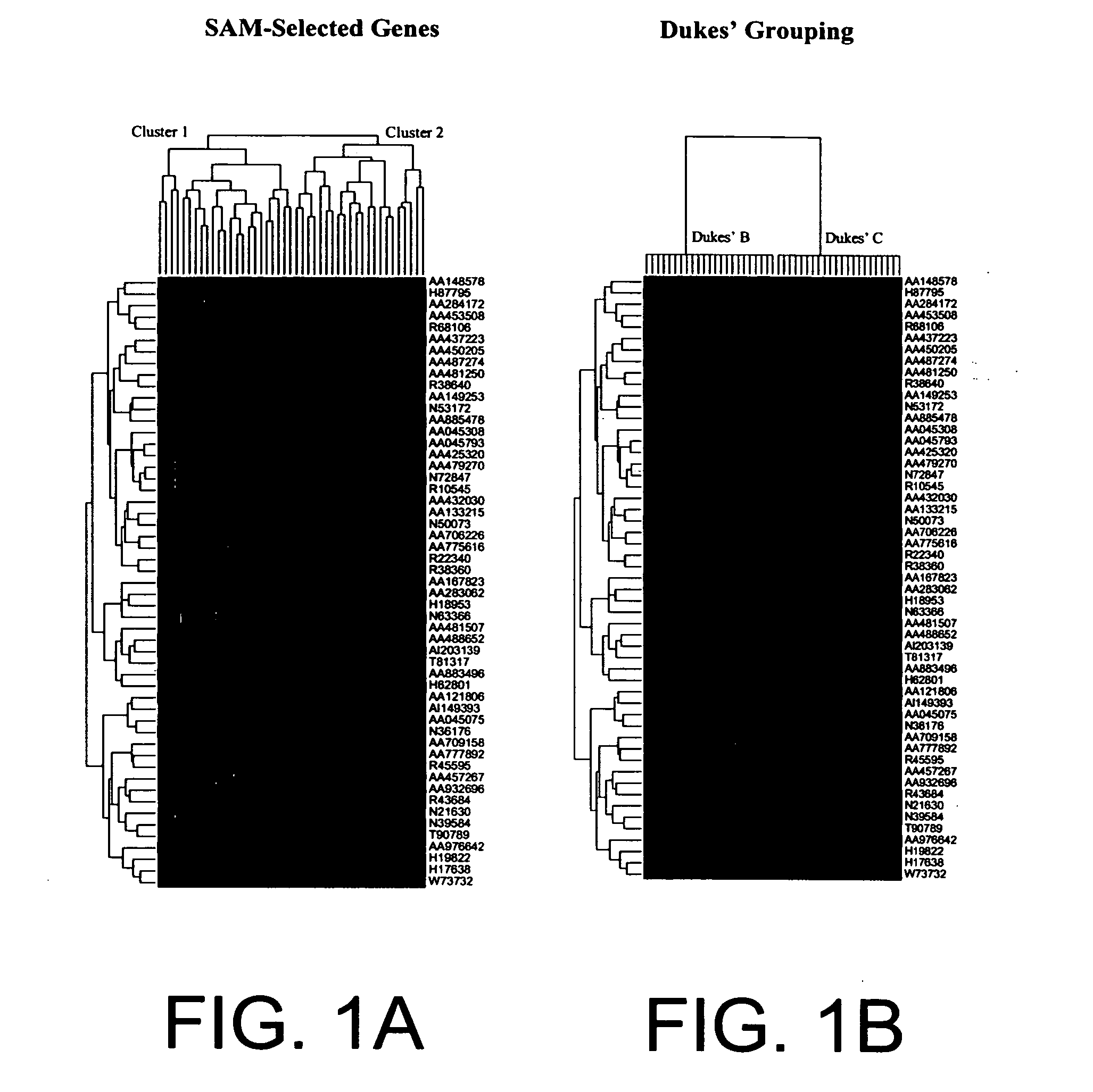
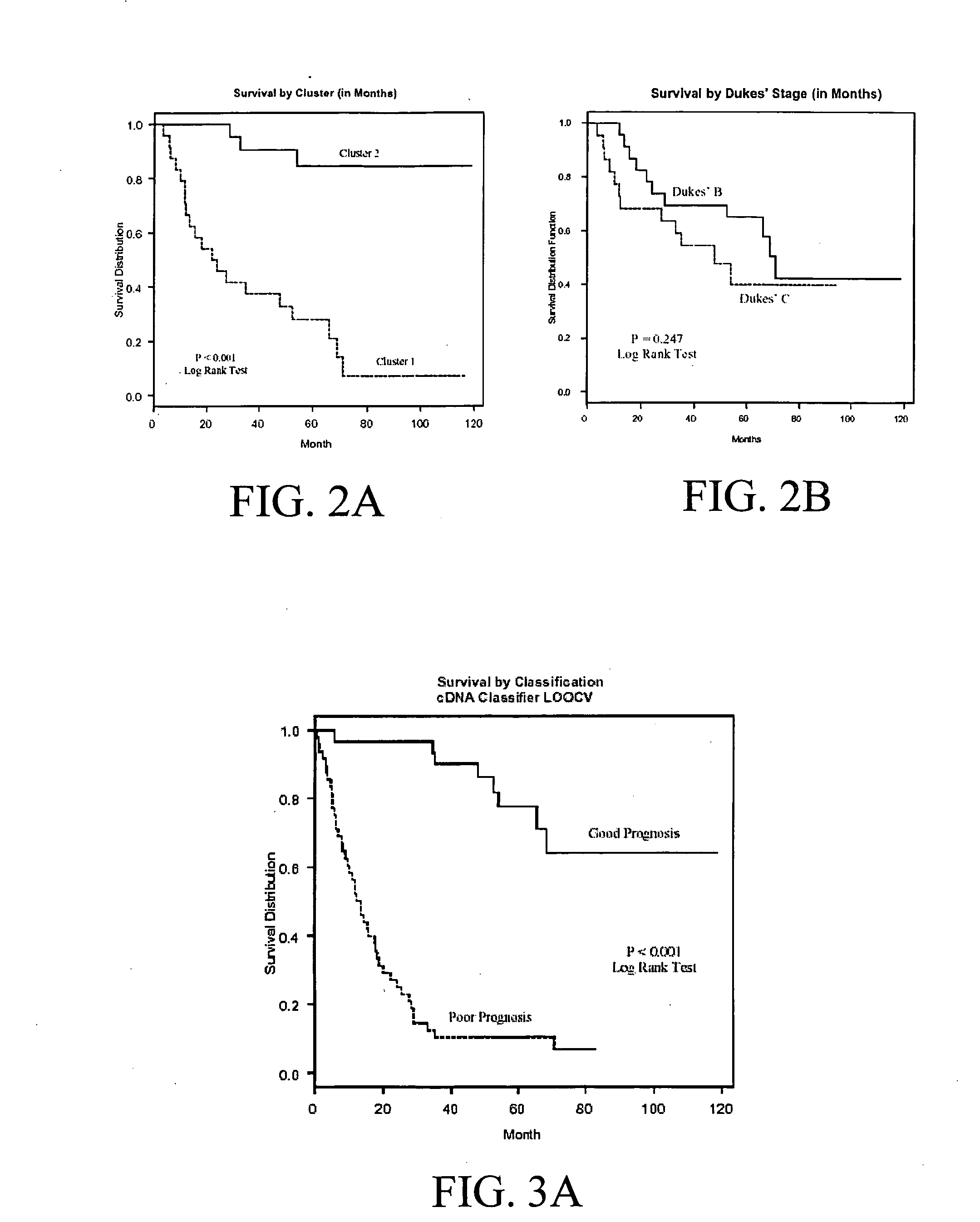
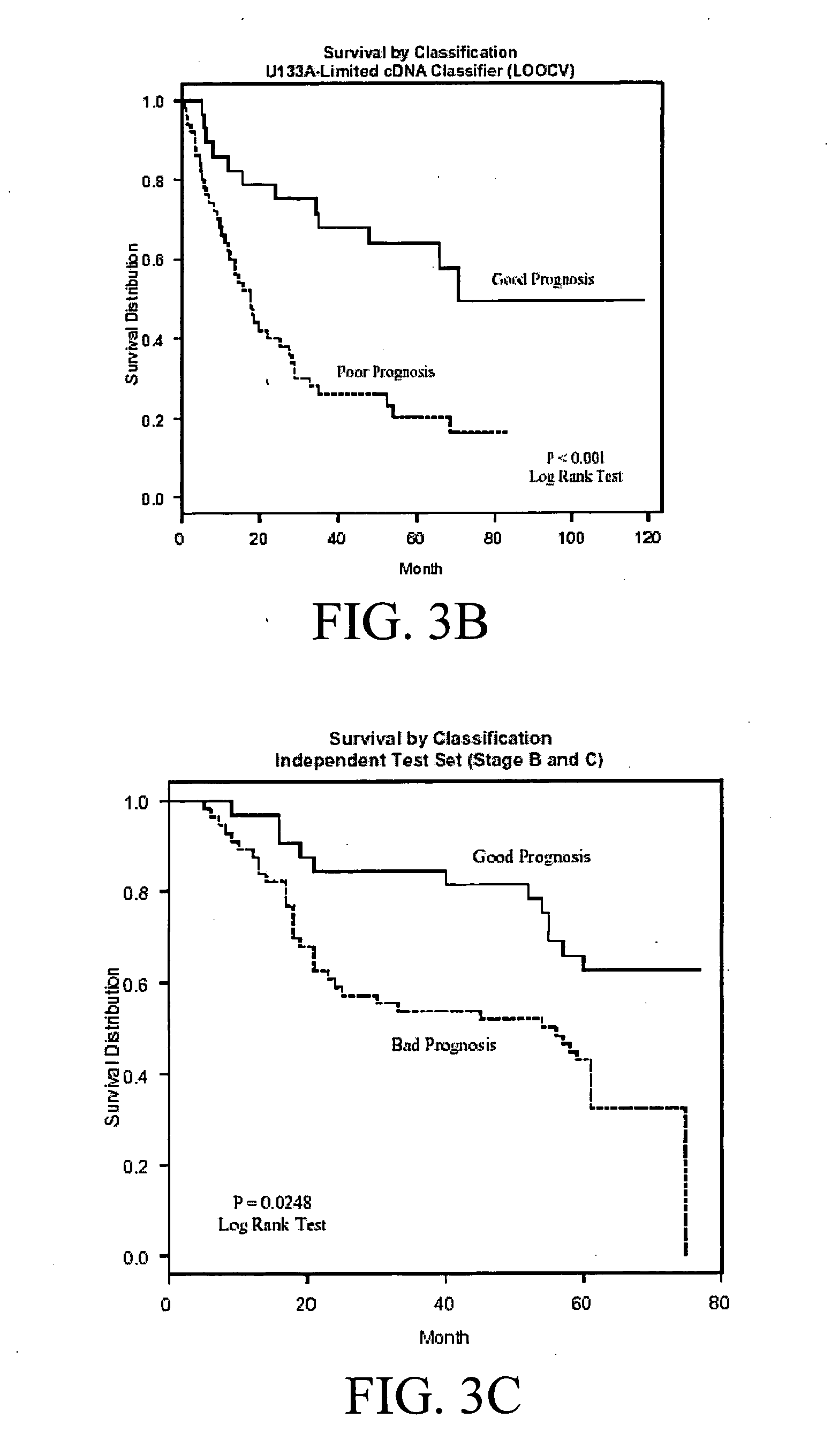
Human Colon Cancer Survival Classifier
Training Set Tumor Samples
[0065] In one embodiment of the subject invention, a colon cancer survival classifier was developed using 78 tumor samples, including 3 adenomas and 75 cancers. Informative frozen colorectal cancer samples were selected from the Moffitt Cancer Center Tumor Bank (Tampa, Fla.) based on evidence for good (survival >36 mo) or poor prognosis (survival <36 mo) from the Tumor Registry. Dukes' stages can include B, C, and D. In this particular embodiment, survival was measured as last contact minus collection date for living patients, or date of death minus collection date for patients who have died.
[0066] In this embodiment, the number of samples per Dukes' stage was as follows: 23 patients with stage B, 22 patients with stage C and 30 patients with stage D disease. Just as adenomas can be included to help train the classifier to recognize good prognosis patients, Dukes D patients with synchronous metastatic disease can be...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com