Gene Signature for the Prediction of Radiation Therapy Response
a gene signature and radiation therapy technology, applied in chemical/physical/physico-chemical processes, instruments, suspensions and porous materials, etc., can solve the problems that the efforts to understand the biological parameters that define intrinsic radiosensitivity have not met the same success, and achieve the effect of increasing or decreasing the radiosensitivity, increasing or decreasing the amount of radiation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
A Radiosensitivity Systems Model Captures Central Regulatory Pathways in Radiation Response
[0134]The model used in the methods described herein was developed in 48 cancer cell lines from the NCI panel of 60 (listed in Table 1). Radiosensitivity measurements (as determined by clonogenic survival at 2 Gy, SF2) were either determined using known methods (Gupta et al., Cancer Res 2001; 61:4278-82; Torres-Roca et al., Cancer Res 2005; 65(16):7169-76) (25 cell lines) or obtained from the literature (23 cell lines). SF2 results for each cell line are presented in Table 1.
TABLE 148 cell lines and measured SF2 values.RecordedCell LineSF2BREAST_HS578T0.79BREAST_MDAMB2310.82COLON_HCT1160.38COLON_HCT150.4COLON_SW6200.62LEUK_CCRFCEM0.185LEUK_HL600.315LEUK_MOLT40.05MELAN_SKMEL20.66NSCLC_A549ATCC0.61NSCLC_H4600.84NSCLC_HOP620.164NSCLC_NCIH230.086OVAR_OVCAR50.408RENAL_SN12C0.62BREAST_BT5490.632BREAST_MCF70.576BREAST_MDAMB4350.1795BREAST_T47D0.52CNS_SF2680.45CNS_SF5390.82CNS_SNB190.43CNS_SNB750.55CN...
example 3
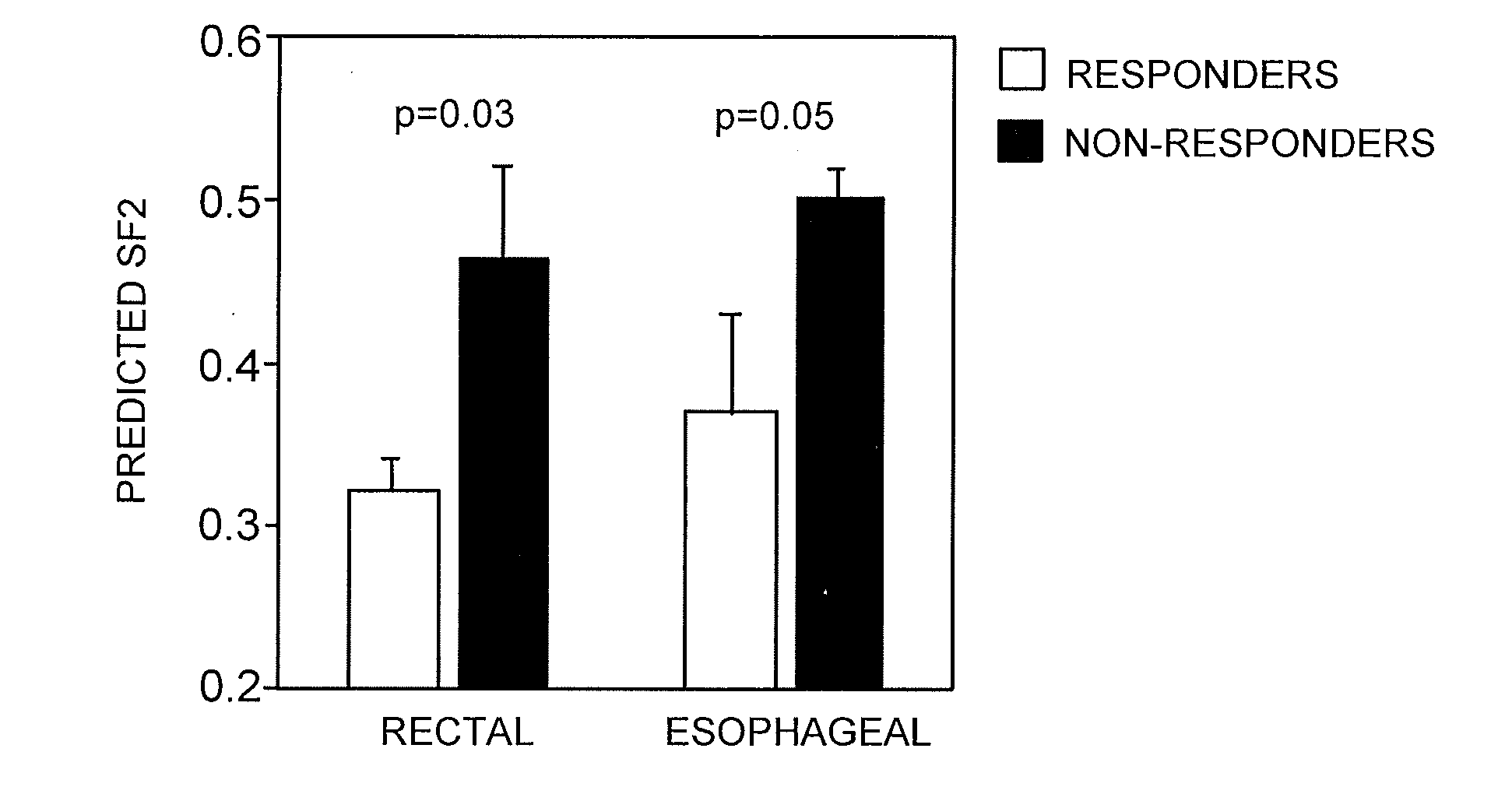
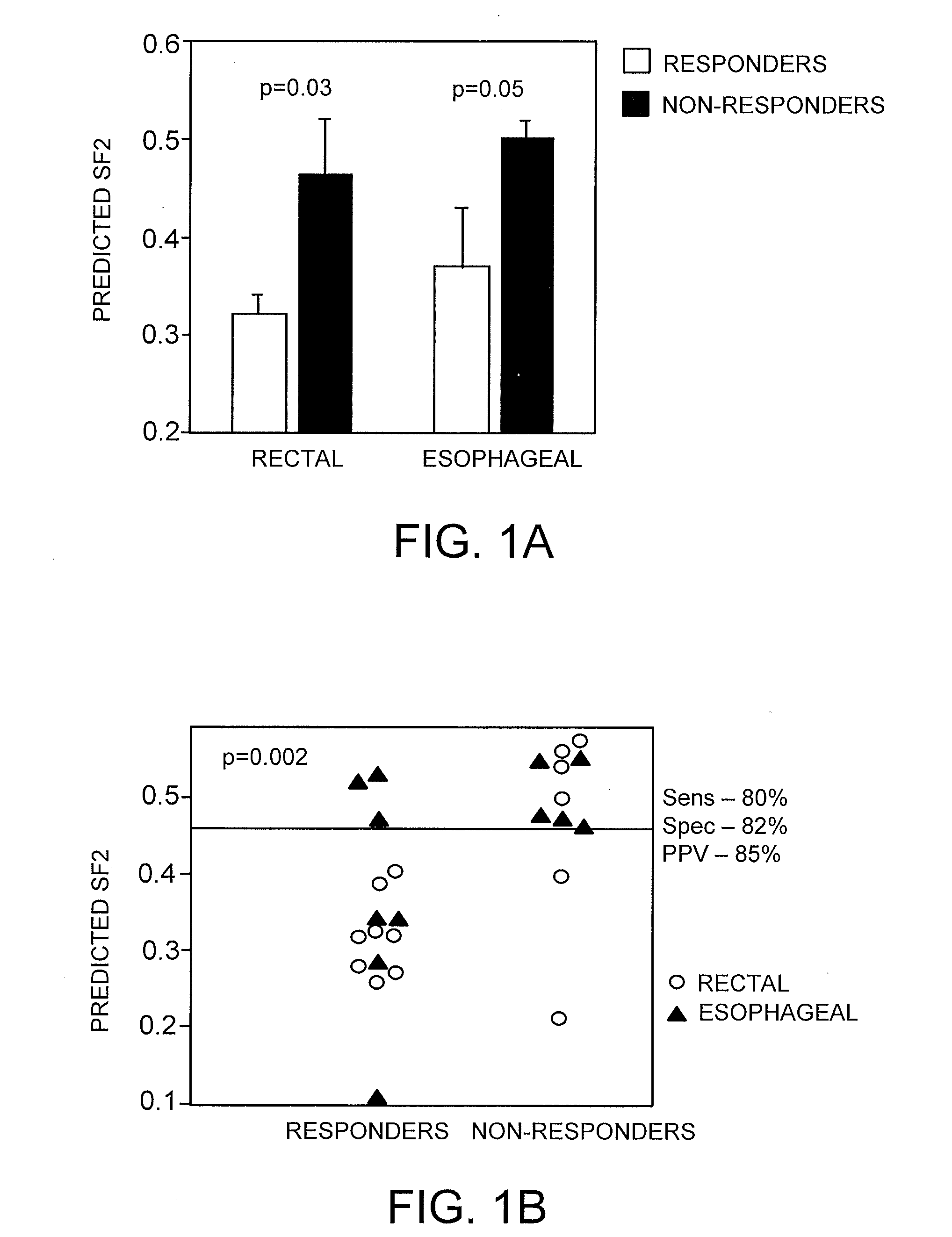
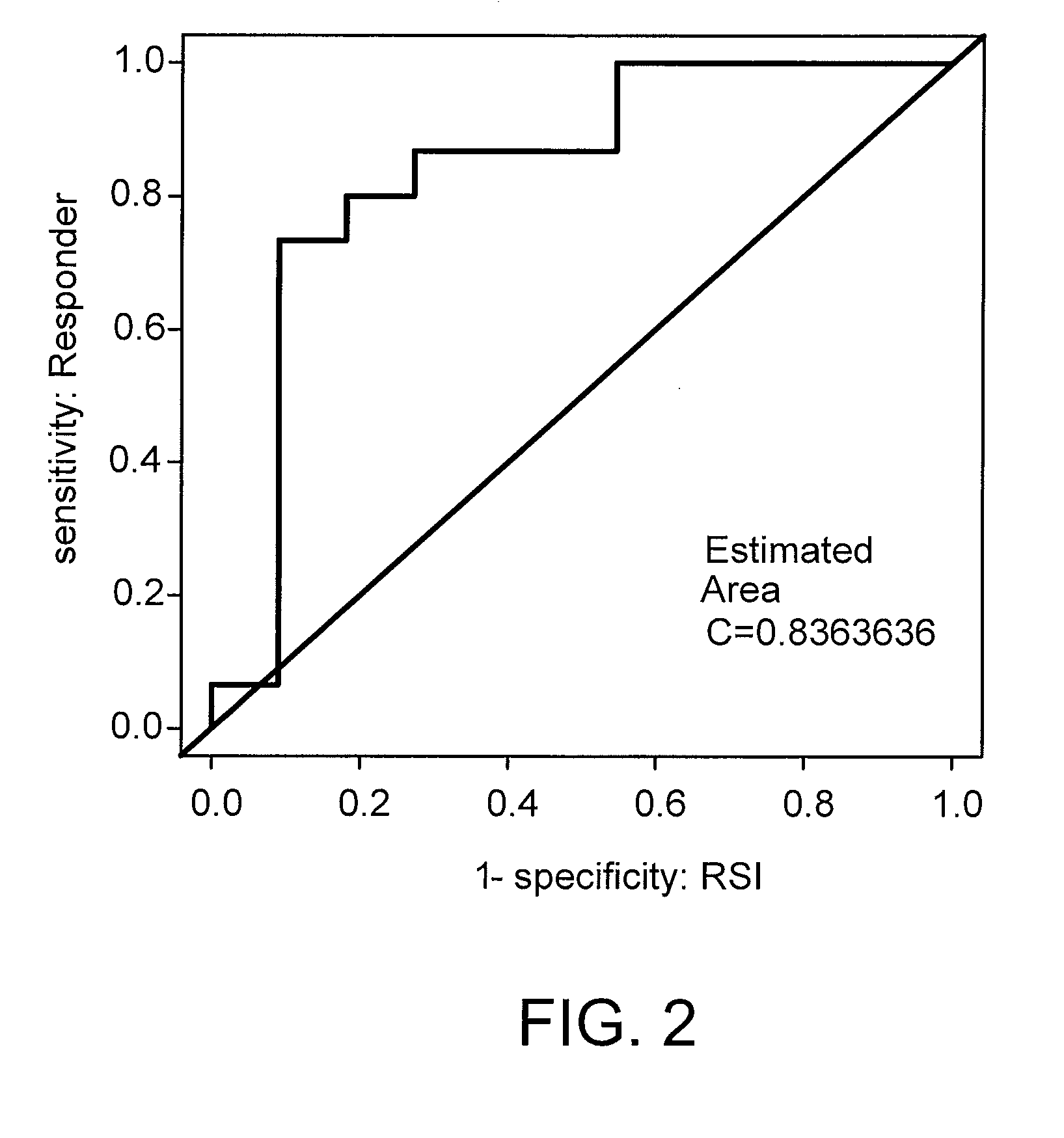
The Radiosensitivity Model Predicts Pathological Response to Chemoradiation in Rectal and Esophageal Cancer
[0153]The regression model developed as described in Examples 1-2 was then applied to similarly rank-ordered patient data to generate a Radiation Sensitivity Index (RSI).
[0154]The model was applied to the prediction of clinical response to concurrent radiochemotherapy in two independent prospectively-collected pilot cohorts of patients with rectal (n=14) and esophageal cancer (n=12). Pathological response was defined by T stage criteria (see methods).
[0155]The Rectal Cancer Cohort consisted of 14 patients enrolled in an IRB-approved prospective Phase 1 trial evaluating escalating doses of oral topotecan as a radiosensitizing agent in patients with rectal cancer. Informed consent was obtained for all patients prior to enrollment. The eligibility criteria included patients with histologically-confirmed rectal cancer with a primary tumor at least 3 cm in size and a clinical stage ...
example 4
The Radiosensitivity Predictive Model is of Prognostic Value in Head and Neck Cancer
[0167]The model was further tested as a prognostic marker in locally-advanced head and neck cancer patients treated with definitive concurrent radiochemotherapy.
[0168]The Head and Neck Cancer Cohort consisted of 92 patients with head and neck cancer treated within prospective randomized Phase II-III trials at The Netherlands Cancer Institute. The majority of tumors were locally-advanced advanced (94% T3 and above, 74% N1 and above). The full clinical details of this cohort were previously published (Pramana et al., Int J Radiat Oncol Biol Phys 2007; 69(5):1544-52). All patients were treated with concurrent radiochemotherapy with cisplatin-based chemotherapy. Total radiation dose was 70Gy in 2Gy daily fractions in all cases. Two different schedules of cisplatin were given: 1. (high dose) 100 mg / m2 IV three times during radiotherapy or 150 mg / m2 given intra-arterially four times during radiotherapy; or...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Therapeutic | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
| Gene expression profile | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com