Application of acetyltransferase osg2 gene and its encoded protein
A technology of acetyltransferase and gene encoding, applied in the field of genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1: Acquisition of transgenic rice material
[0034] 1. Obtainment of transgenic mutant plants
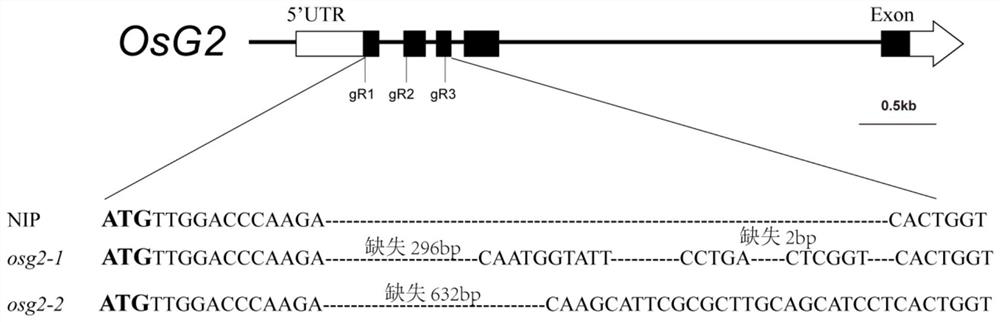
[0035] The plant materials used in this study include wild-type Nipponbare rice and two mutant plants, and the two mutant plants are named osg2-1 and osg2-2 respectively. After gene editing by CRISPRCas9 technology, the result of gene editing is to produce two allelic mutants with different mutation sites. The editing method is as follows: figure 1 shown.
[0036] The full length of OsG2 gene is 4256bp, with figure 1 Gene structure diagram shown. Among them, NIP represents wild-type Nipponbare rice, which is used as a control; in order to construct mutant plants of OsG2 gene, CRISPR-Cas9 gene editing technology was used to create them. The main methods are as follows:
[0037] The transformed plants of the T0 generation of rice were detected by PCR sequencing, and the double-stranded homozygous edited mutants were screened and named osg2-1 and osg2-2, respectively....
Embodiment 2
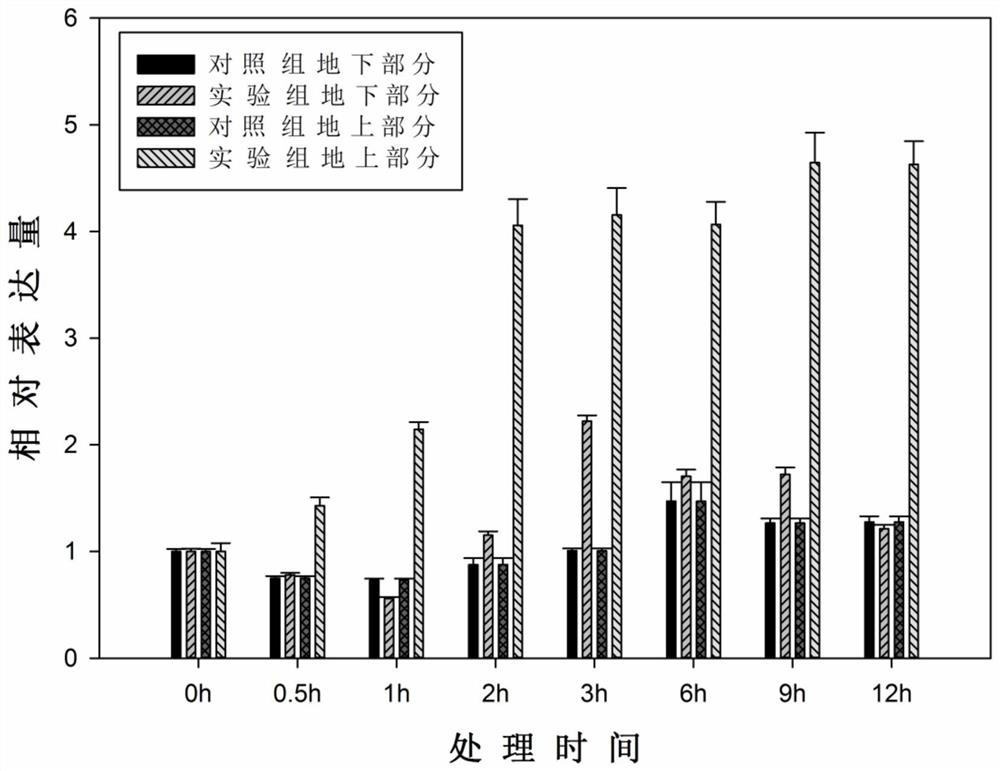
[0058] Embodiment 2: the response situation of analysis OsG2 of salt stress treatment
[0059] First, the wild-type Nipponbare was used for salt stress treatment, and the response of OsG2 was analyzed. The specific process is as follows: select wild-type Nipponbare rice seeds in good development condition, put them into a petri dish containing a small amount of sterile water without shelling, and lay a layer of absorbent paper on the bottom of the petri dish. Put the petri dish containing the seeds in a 30-35 ℃ incubator for two days, and then transfer the seeds to an incubator with a temperature of about 25 ℃ after the seeds are white and broken to achieve the purpose of promoting germination and rooting. It should be noted that in the process of seed soaking and germination, it is necessary to diligently replace the sterile water in the petri dish to ensure the normal respiration and growth of the seeds.
[0060] The rice seedlings with the same growth status after germinat...
Embodiment 3
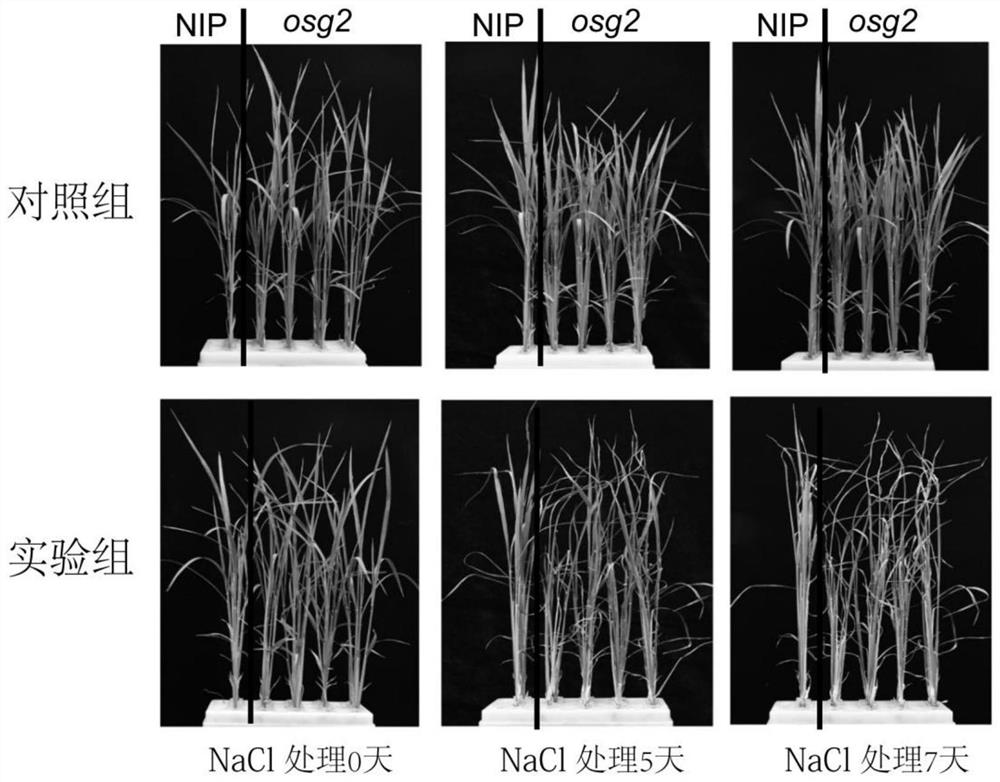
[0068] Example 3: Detection of salt sensitivity of transgenic mutant plants
[0069] Experimental materials: wild-type Nipponbare, OsG2 gene allelic mutant
[0070] Treatment method: The germination method of rice seeds is as described in Example 2. The wild-type Nipponbare and mutant seedlings with the same growth state were selected, transferred to Hoagland nutrient solution containing 150 mM NaCl, and treated for 0 days, 5 days and 7 days respectively. The morphological changes of leaves in the control group and the experimental group were recorded by phenotype. like image 3 shown, the mutant plants were more sensitive to salt stress than the wild-type Nipponbare.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com