Protein-protein interaction site prediction method based on deep map convolutional network
An interaction site, convolutional network technology, applied in neural learning methods, biological neural network models, instruments, etc., can solve problems such as inability to extract amino acid information, inability to effectively simulate complex relationships of amino acids, and achieve the effect of improving accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
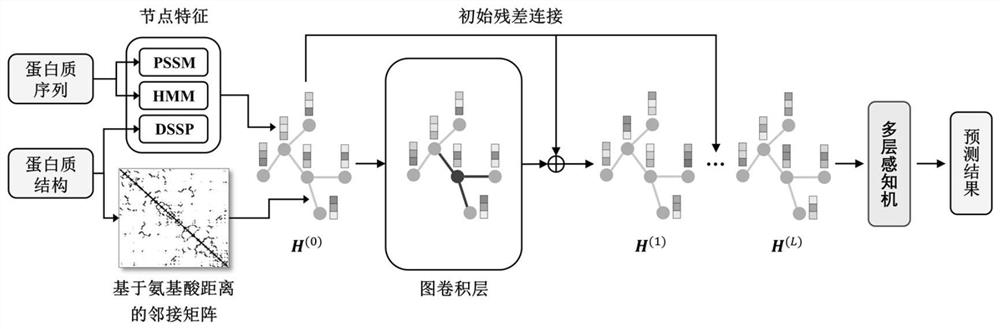
[0048] like figure 1 As shown, a protein-protein interaction site prediction method based on a deep graph convolutional network, the method includes the following steps:
[0049] S1: According to the sequence and structural information of the protein, extract the node feature matrix and the adjacency matrix containing edge information to form a protein graph representation together;
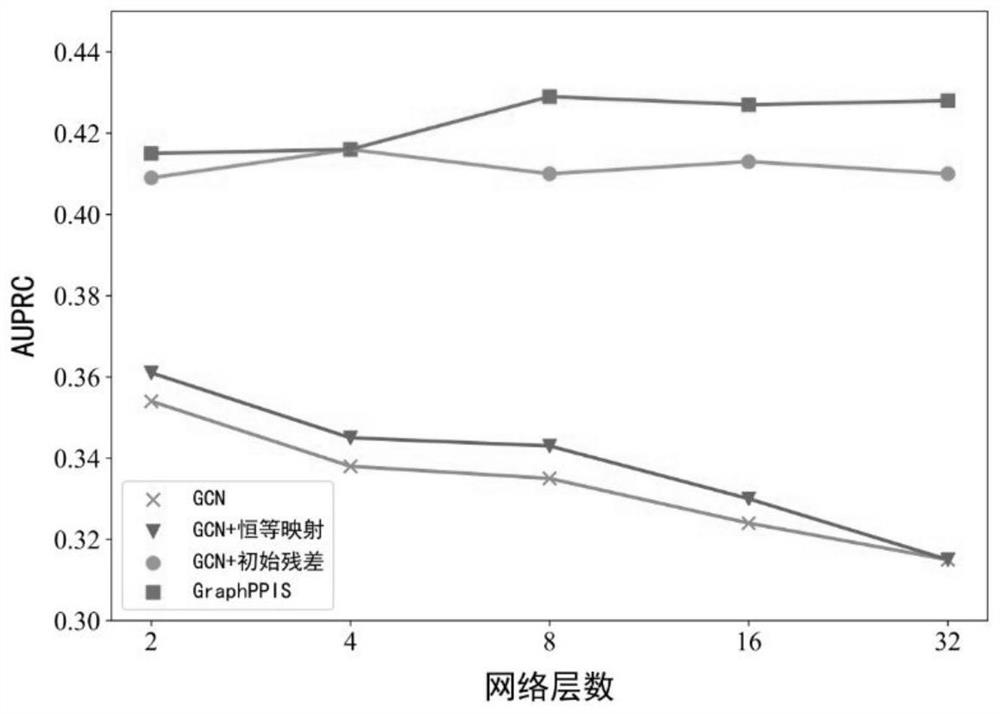
[0050]S2: Use deep graph convolution based on initial residual and identity mapping to capture the characteristics of high-order spatially adjacent amino acids; and input a multi-layer perceptron at the output of the last graph convolution layer of deep graph convolution, Realize the final prediction of the protein interaction probability of each amino acid, and complete the construction of a deep graph convolutional neural network;
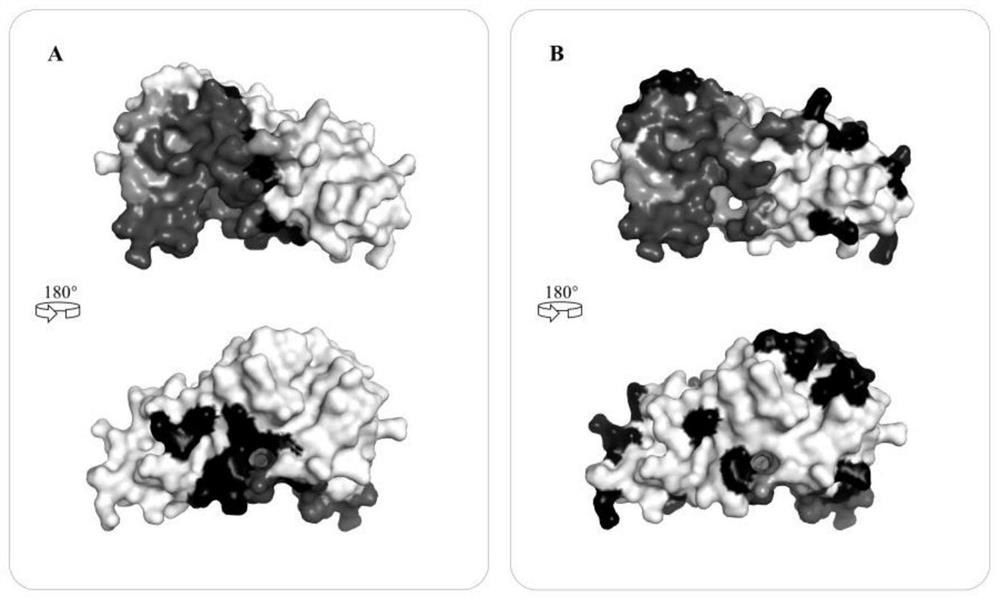
[0051] S3: extract the training data through step S1 to obtain the protein graph representation, and use the five-fold cross-validation method to train the deep gr...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap