SNP (Single Nucleotide Polymorphism) molecular marker located on 12 # chromosome of pig and related to piglet malformation number and application of SNP molecular marker
A molecular marker and chromosome technology, applied in recombinant DNA technology, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problem of inability to fundamentally solve the genetic defects of pigs, restrict the rapid transmission of excellent genes, and eliminate individuals. Achieve the effect of increasing core competitiveness, reducing the number of deformed piglets, and ensuring corporate profits
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] (1) The extraction method of pure Duroc pig ear-like tissue DNA was extracted from the whole genome DNA according to the marked phenol-chloroform method. The DNA quality and concentration of the pure Duroc population were tested with a Nanodrop-ND1000 spectrophotometer. The ratio of A260 / 280 is 1.8-2.0, and the ratio of A260 / 230 is 1.7-1.9. Finally, the qualified DNA samples were uniformly diluted to 50 ng / μl. Then mix 6 μL of the extracted DNA sample to be tested with 2 μL Loading Buffer, load the sample on agarose gel with a mass-volume ratio of 1%, electrophoresis at 150V for 25 min, and perform electrophoresis under UV spectrophotometer and gel imaging equipment Observe and take pictures to observe the integrity of DNA. After passing the DNA integrity test, the genotype test was performed.
[0035] (2) Detection of 50K SNP genotype of the whole pig genome: DNA samples were sent to Neogen Biotechnology (Shanghai) Co., Ltd., and the genotype of the pig whole genome...
Embodiment 2
[0042] Example 2 Detection of SNP molecular markers
[0043] (1) The target fragment containing the SNP site significantly related to the number of deformities in offspring of Duroc pigs is a 224bp nucleotide sequence in chromosome 12, and the upstream and downstream primers for sequence amplification are primer-F and primer-R , its nucleic acid sequence is as follows:
[0044] Upstream primer primer-F: 5'-TGCCTGGCCTCTCACTGTCACAC-3';
[0045] Downstream primer primer-R: 5'-ACTCCTCTTTGTCCCCGGCTT-3';
[0046] (2) PCR amplification system and condition setting
[0047] Configure a 10 μL system, including 1.0 μL DNA sample, 0.3 μL upstream primer, 0.3 μL downstream primer, 5.0 μL PCR StarMix, ddH 2 O 3.4 μL, PCR conditions were pre-denaturation at 94°C for 2 min, denaturation at 94°C for 30 s, annealing at 64°C for 30 s, extension at 72°C for 15 s, a total of 35 cycles, and a final extension at 72°C for 5 min.
[0048] (3) DNA sequence sequencing identification: Sequence seque...
Embodiment 3
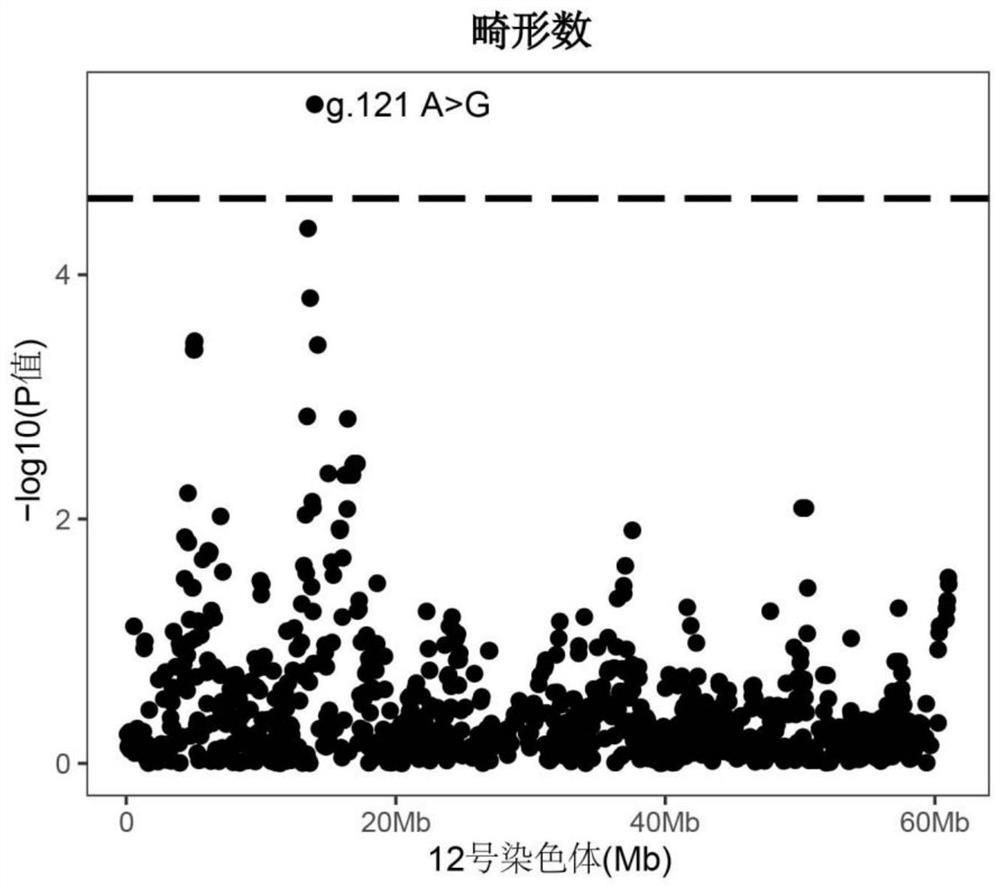
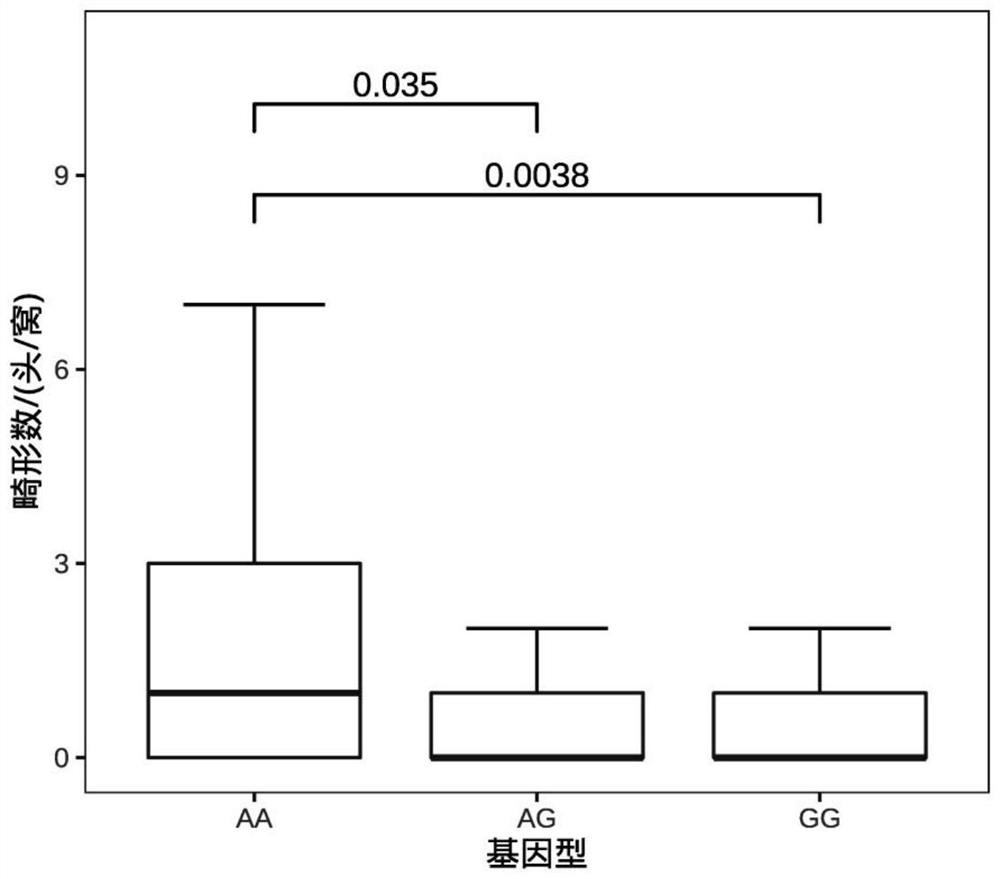
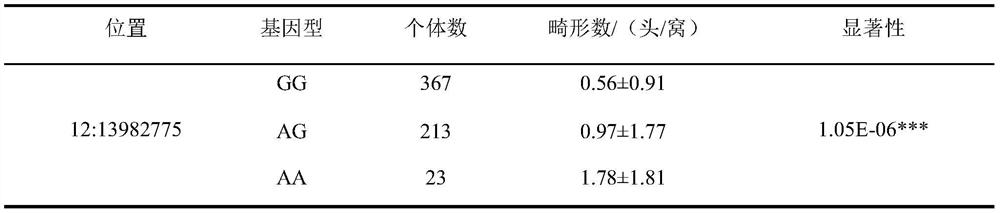
[0051] Example 3 SNP site g.121A>G effect analysis of molecular markers
[0052] According to Table 1, figure 2 It can be seen that the molecular marker SNP site g.121A>G (the 121st nucleotide in SEQ NO.1) has a very significant correlation with the number of piglet deformities. The number of deformed piglets of GG and AG type individuals was less than that of AA type individuals, and the number of deformed piglets of GG type individuals was 1.22 less than that of AA type individuals. Therefore, keeping the breeding pigs of GG and AG genotypes in breeding to gradually increase the frequency of the allele G at this site can significantly reduce the number of piglet deformities and bring greater economic benefits to the enterprise.
[0053] The present invention detects the 121st base mutation site in the sequence of SEQ ID NO: 1, and preliminarily conducts a correlation analysis between its genotype and the occurrence of piglet deformities, and provides a new molecular marker...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com