Primer pair probe combination product and its kit and detection method for detecting mitochondrial 3243a>g mutation
A combination product and detection probe technology, applied in the field of genes, can solve the problems of not being able to provide guidance to doctors in time, high cost of chip method, and long time period, and achieve intuitive and easy interpretation of results, short detection time consumption, and low cost Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
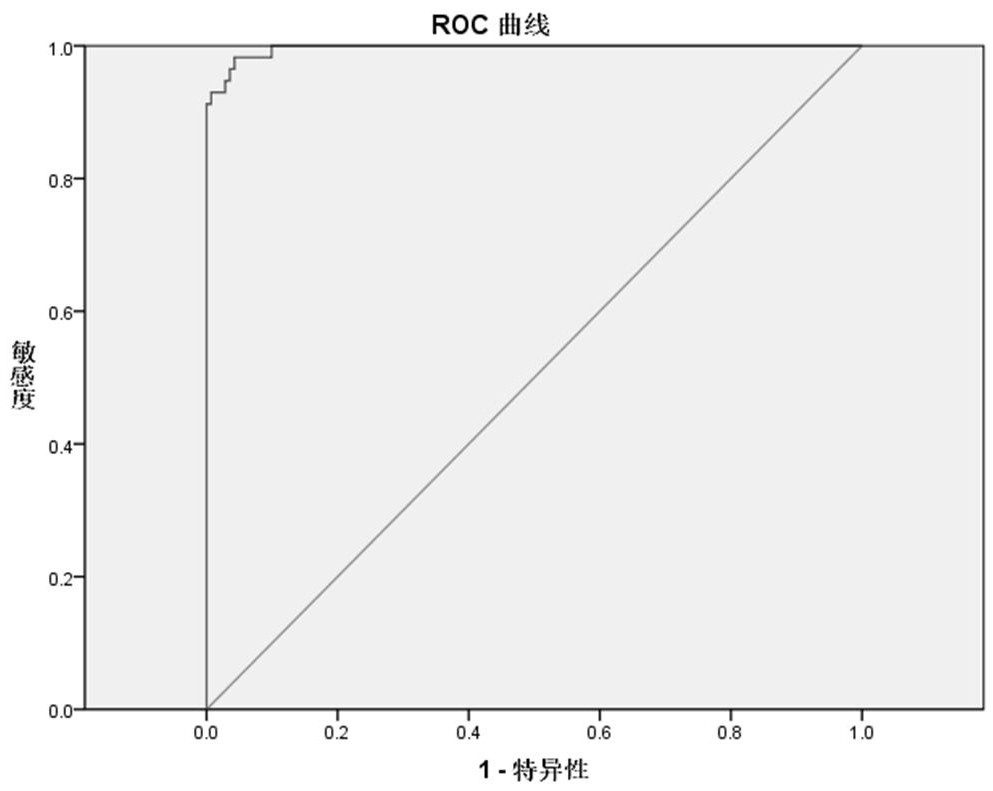
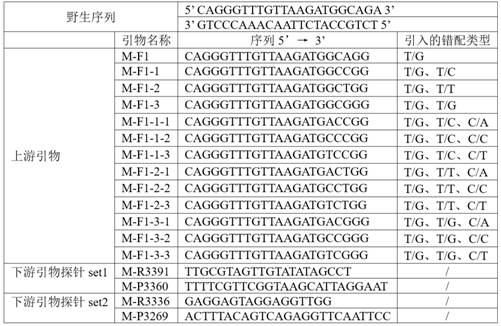
[0058] Example 1 Screening of primers and probes
[0059] Step 1: m. 3243A>G primer and probe design and selection
[0060] Find the sequence near the mitochondrial site 3243: Search the mitochondrial genome sequence information (NC_012920.1) on NCBI, intercept the sequence of 300~500bp upstream and downstream of the 3243 site for primer probe design, and the underlined one is m.3243 A> G-site.
[0061] GTACGAAAGGACAAGAGAAATAAGGCCTACTTCACAAAGCGCCTTCCCCCGTAAATGATATCATCTCAACTTAGTATTATACCCACACCCACCCAAGAACAGGGTTTGTTAAGATGGCAG A GCCCGGTAATCGCATAAAACTTAAAACTTTACAGTCAGAGGTTCAATTCCTCTTCTTAACAACATACCCATGGCCAACCTCCTACTCCTCATTGTACCCATTCTAATCGCAATGGCATTCCTAATGCTTACCGAACGAAAAATTCTAGGCTATATACAACTACGCAAAGGCCCCAACGTTGTAGGCCCCTACGGGCTACTACAACCCTTCGCTGACGCCATAAAACTCTTCACCAAAGAGCCCCTAAAACCCGCCACATCTACCATCACCCTCTACATCACCGCCCCGACCTTAGCTCTCACCATCGCTCTTCTACTA
[0062] With the aid of primer and probe design software oligo 7 and Primer Premier 5.0 software, 13 upstream primers and 2 sets of downstr...
Embodiment 2
[0084] Embodiment 2: Optimization of reaction system and reaction program
[0085] The optimal reaction system was established through the study of different Mix usage amounts, different primers, probe concentrations and amounts, and different amounts of DNA templates; the optimal reaction program was established through the research of different annealing temperatures, times, and cycle numbers. The reaction system and reaction program are shown in Table 8-Table 11.
[0086] Specifically, the mitochondrial gene 3243A>G mutation detection kit (fluorescent PCR method) includes the following steps:
[0087] S1: Nucleic acid extraction;
[0088] S2: Reaction solution configuration;
[0089] S3: add sample;
[0090] S4: qPCR amplification;
[0091] S5: Data analysis.
[0092] Step S1 is specifically: vortex the centrifuge tube containing the oral swab for 1 min to completely elute the exfoliated cells in the oral cavity, and discard the swab. Take 0.2ml of oral exfoliated cel...
Embodiment 3
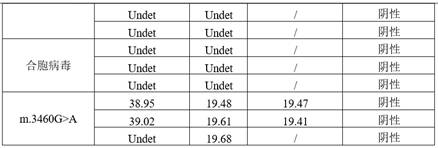
[0115] Embodiment 3: the research of positive judgment value
[0116] Step 1: Determine the reference value of the internal reference gene
[0117] When the concentration of added DNA is excessive, it is easy to generate false positive signals. When the concentration of DNA is insufficient or PCR has inhibitors, false negative signals are prone to occur. Therefore, it is necessary to use the internal reference gene amplification to monitor the quality of the added DNA template and to monitor each DNA amplification in each reaction well.
[0118] The DNA of 60 oral swab samples was randomly selected for 10-fold serial dilution to 0.1ng / μL for detection, and the reference range of Ct value was calculated. The results show that when the DNA concentration is 0.1ng / μL, the Ct value is distributed between 23 and 26, the average value is 24.34, and the standard deviation SD is 0.75. The reference range of the Ct value of the internal reference is the average value plus 2 times the s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com