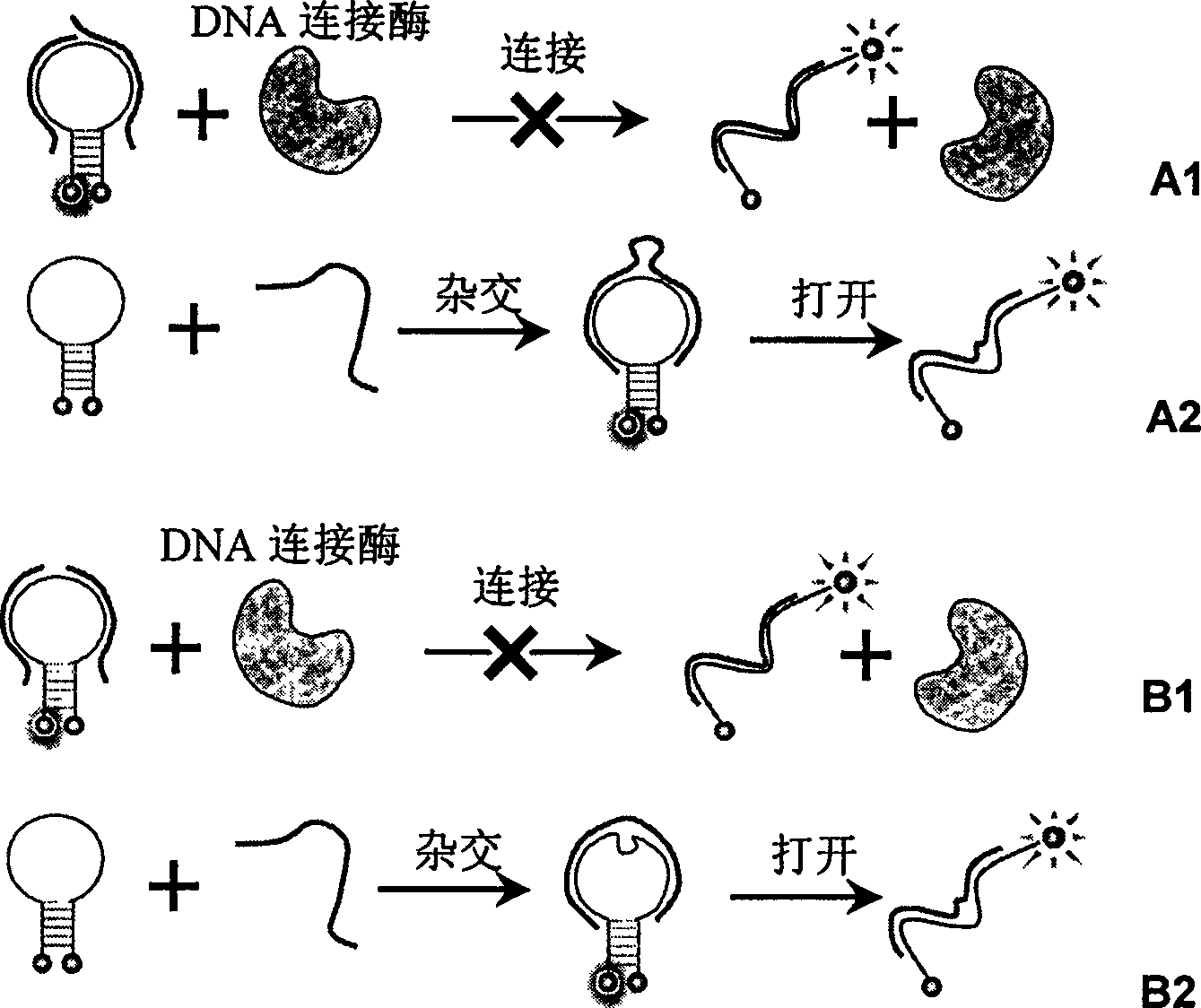
Real time method for detecting nucleic acid ligase reaction and nucleic acid ligase chain reaction
A ligase reaction, real-time detection technology, applied in the field of molecular biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
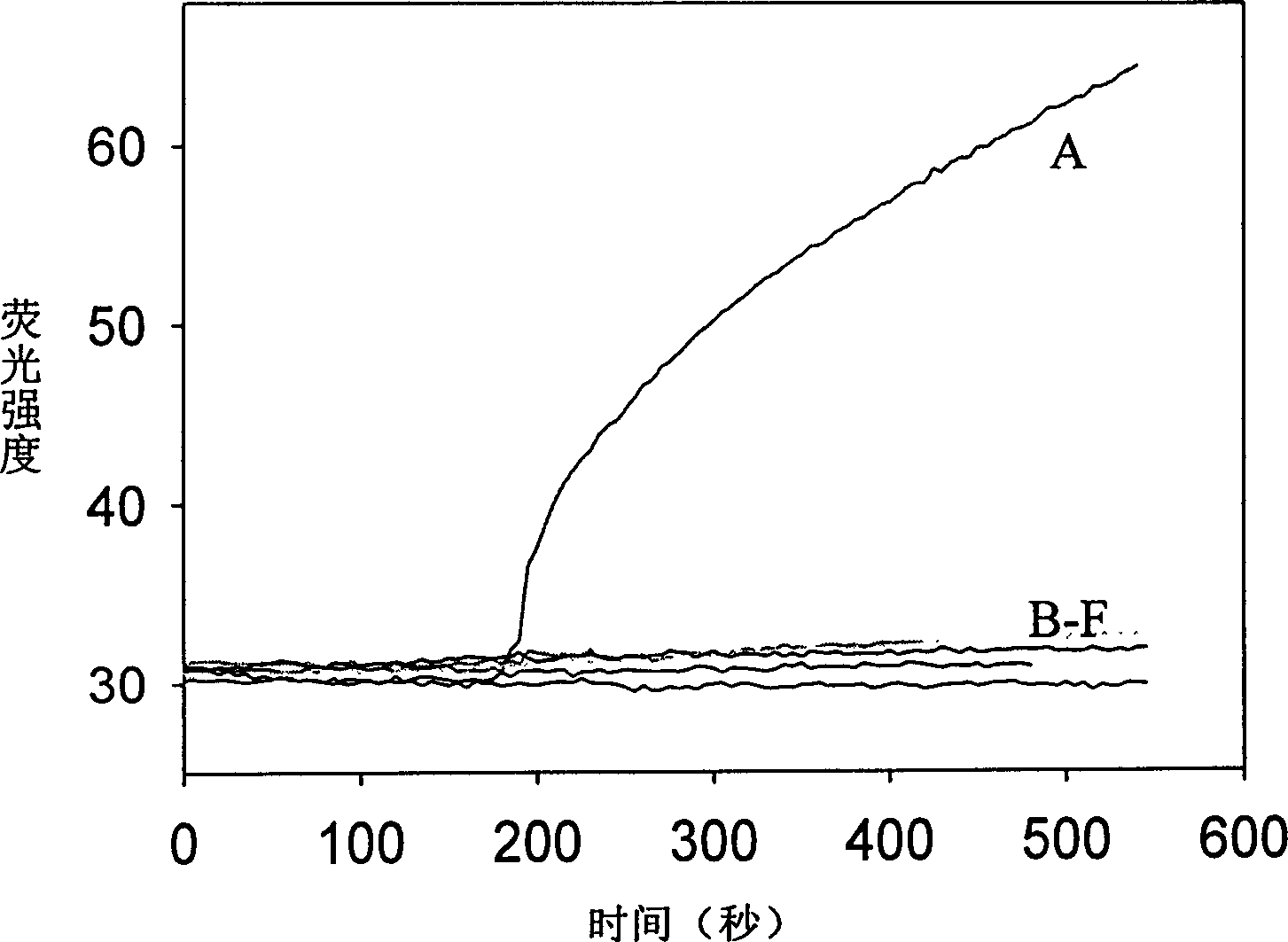
[0019] Embodiment 1 (verification experiment of detection method principle)
[0020] Six samples were prepared in the experiment, in 200uL sample buffer (including 20mM Tris-HCl (pH=7.6), 25mM KAc, 10mM Mg(Ac) 2 , 10mM DTT, 1mM NAD, 0.1% Triton-X100) and MB with a final concentration of 200nM, in addition, different DNA primers were added to each sample: (A) Primer1+Primer4; (B) Primer1+Primer5; (C) Primer2 +Primer5; (D) Primer1+Primer6; (E) Primer3+Primer4; (F) Primer3+Primer6; where the final concentration of Primer1 to Primer6 is 200 nM. The sample was placed in a F2500 fluorometer, and the sample temperature was kept constant at 45 degrees Celsius. Then the fluorescence intensity of the sample was detected with the parameters of excitation wavelength 497nm and emission wavelength 521nm. After the fluorescence intensity is stable, add 10U of Taq ligase, monitor and record the fluorescence intensity of the sample in real time, the results are as follows: figure 2 shown. ...
Embodiment 2
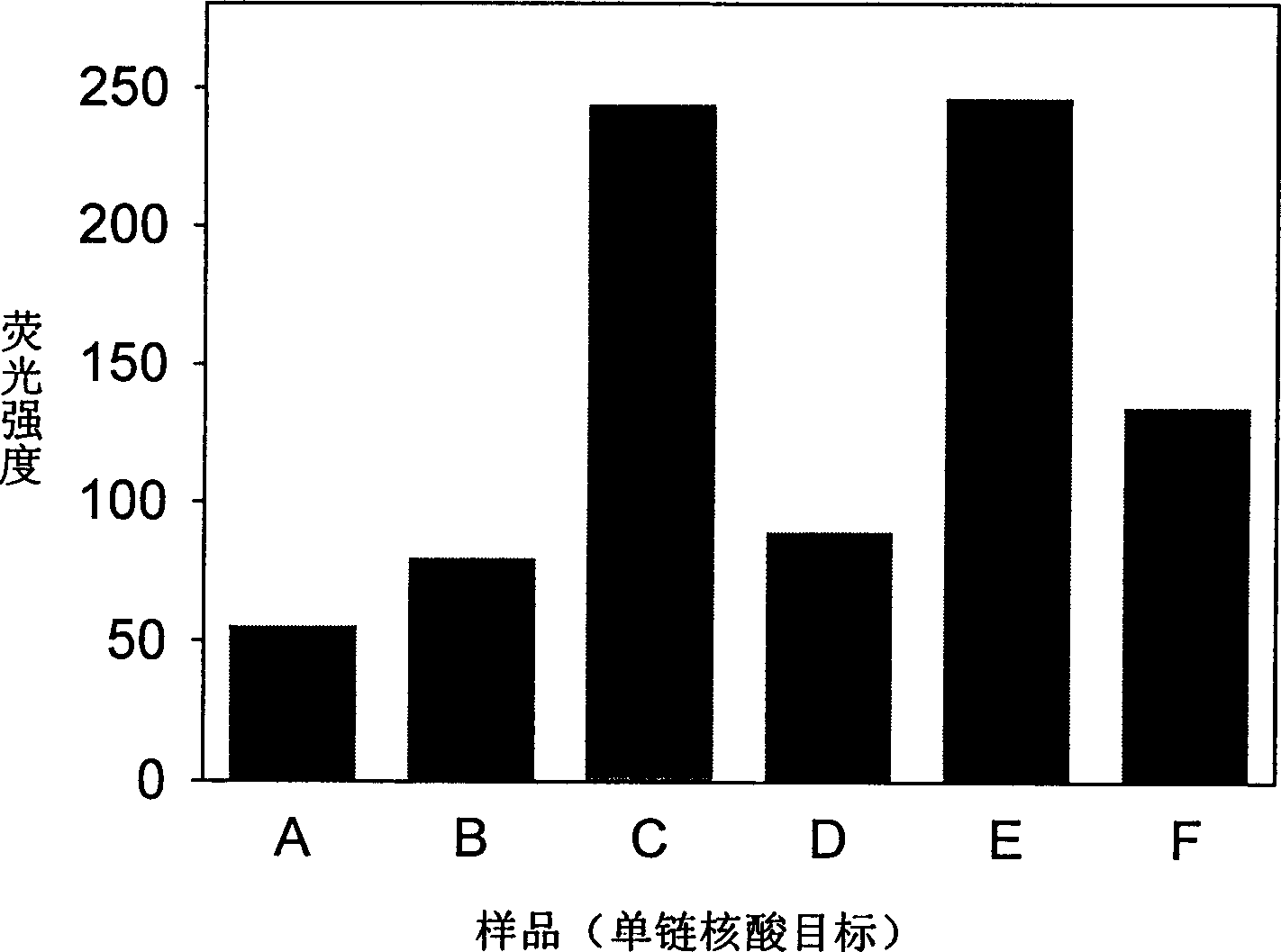
[0027] Embodiment 2 (real-time LDR method is to the analysis of single-stranded nucleic acid object)
[0028] Six samples were prepared in the experiment, in 200uL sample buffer (including 20mM Tris-HCl (pH=7.6), 25mM KAc, 10mM Mg(Ac) 2 , 10mM DTT, 1mM NAD and 0.1% Triton-X100) were added with 30U Taq ligase and MB with a final concentration of 200nM, except for one MB control sample A without DNA samples, other samples contained (B) PrimerA +PrimerB; (C) MutantDNA1+PrimerA+PrimerB; (D) NormalDNA1+PrimerA+PrimerB; (E) MutantDNA1+PrimerC+PrimerD; (F) NormalDNA1+PrimerC+PrimerD; is 200nM, and the concentration of template DNA is 10nM. Put these 5 samples in a PCR machine, set the program for LDR temperature cycle, stay at 45 degrees Celsius for 5 minutes for ligation amplification, raise the temperature to 80 degrees Celsius for 3 minutes to melt the DNA, and do 25 cycles in total. Then, the fluorescence intensity of the sample was detected with a F2500 fluorescence instrument...
Embodiment 3
[0030] Embodiment 3 (real-time LDR method is to the analysis of double-stranded nucleic acid object)
[0031]In the experiment, 6 samples were prepared, and 30U of Taq ligase and MB with a final concentration of 200nM were added to 200uL sample buffer (same as Example 2). Except for the control sample A of one MB without adding other DNA samples, other samples (B) PrimerA+PrimerB; (C) MutantDNA1+MutantDNA2+PrimerA+PrimerB; (D) NormalDNA1+NormalDNA2+PrimerA+PrimerB; (E) MutantDNA1+MutantDNA2+PrimerC+PrimerD; (F) NormalDNA1+NormalDNA2+ PrimerC+PrimerD; wherein the final concentration of the amplification primers PrimerA to PrimerD is 200nM, and the concentration of the template DNA is 10nM. Put these 5 samples in a PCR machine, set the program for LDR temperature cycle, stay at 45 degrees Celsius for 5 minutes for ligation amplification, raise the temperature to 80 degrees Celsius for 3 minutes to melt the DNA, and do 25 cycles in total. Then, the fluorescence intensity of the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com