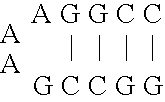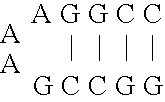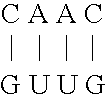Bioreactive allosteric polynucleotides
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0054] As mentioned above, natural ribozymes (8) and ribozymes that have been isolated by in vitro selection are not known to operate as allosteric enzymes (6). This example illustrates allosteric ribozymes.
[0055] Using simple rational design concepts, aptamer domains with hammerhead self-cleaving ribozymes (13) were joined in a modular fashion, to create a series of catalytic RNAs that are amenable to both positive and negative allosteric control by small-molecule effectors. Initial efforts were focused on the 40-nucleotide ATP-binding aptamer, termed ‘ATP-40-1′, that was described by Sassanfar and Szostak (35). This motif shows a specific affinity for adenosine 5′ triphosphate (ATP; KD ˜10 μM) and adenosine, but has no detected affinity for a variety of ATP analogues including 2′-deoxyadenosine 5′ triphosphate (dATP) or the remaining three natural ribonucleoside triphosphates. The aptamer also undergoes a significant conformational change upon ligand binding, as determined by che...
example 2
[0065] The isolation by in vitro selection of two distinct classes of self-cleaving DNAs from a pool of random-sequence oligonucleotides are reported in this example. Individual catalysts from ‘class I’ require both Cu2+ and ascorbate to mediate oxidative self-cleavage. Individual catalysts from class II were found to operate with copper as the sole cofactor. Further optimization of a class II individual by in vitro selection yielded new catalytic DNAs that facilitate Cu2+-dependent self-cleavage with rate a enhancement that exceed 1 million fold relative to the uncatalyzed rate of DNA cleavage.
[0066] DNA is more susceptible to scission via depurination / β-elimination or via oxidative mechanisms than by hydrolysis (27). To begin a comprehensive search for artificial DNA-cleaving DNA enzymes, DNAs that facilitate self-cleavage by a redox-dependent mechanism were screened for. Cleavage of DNA by chelates of redox-active metals (e.g., Fe3+, Cu2+) in the presence of a reducing agent is ...
example 3
[0087] This example describes a DNA structure that can cleave single-stranded DNA substrates in the presence of ionic copper. This deoxyribozyme can self-cleave, or it can operate as a bimolecular complex that simultaneously makes use of duplex and triplex interactions to bind and cleave separate DNA substrates. DNA strand scission proceeds with a kobs of 0.2 min−1, a rate that is ˜1012-fold faster than the uncatalyzed rate of DNA phosphoester hydrolysis. The duplex and triplex recognition domains can be altered, making possible the targeted cleavage of single-stranded DNAs with different nucleotide sequences. Several small synthetic DNAs were made to function as simple ‘restriction enzymes’ for the site-specific cleavage of single-stranded DNA.
[0088] A Minimal Cu2+-Dependent Self-cleaving DNA. In Example 2, a variety of self-cleaving DNAs were isolated by in vitro selection from a pool of random-sequence DNAs. Most individual DNAs that were isolated after eight rounds (G8) of sele...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Mass | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Composition | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com