Anti-viral compositions containing pikfyve inhibitors and use thereof
a technology of antiviral compositions and inhibitors, which is applied in the field of antiviral compositions, can solve the problems of no approved therapies, difficult diagnosis confirmation in the field, and general limitations in evd care, so as to prevent or ameliorate a cytokine storm, reduce the viral load of the subject, and reduce the rate of viral replication
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
is a Highly Selective Binder of PIKfyve Kinase
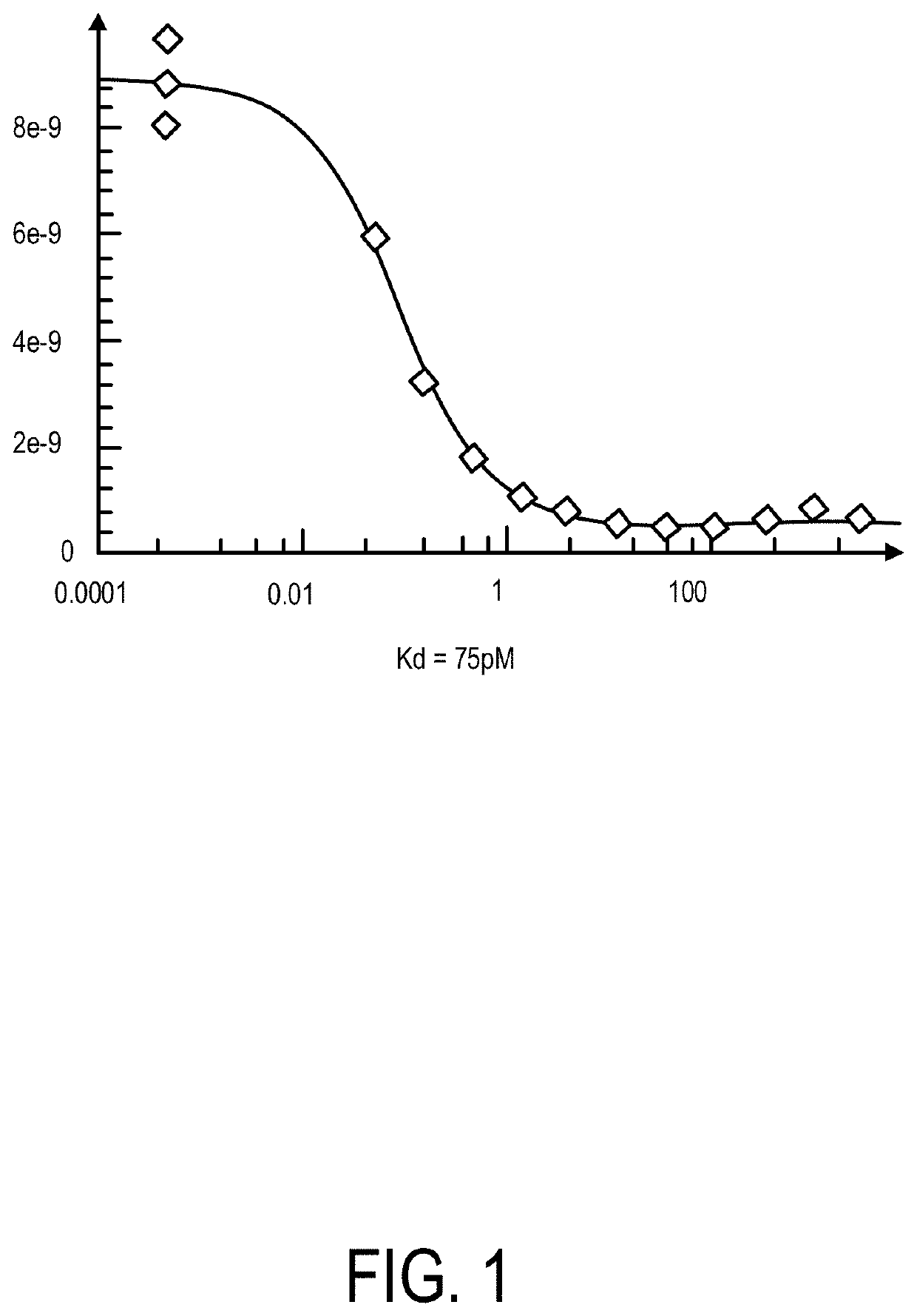
[0098]Protein kinase profiling of apilimod was conducted to identify kinase targets (DiscoveRx, Fremont, Calif.). A dissociation constant (Kd) study was performed using apilimod at increasing concentrations (0.05-3000 nM) against PIKfyve, a known target of apilimod. The experiment was performed in duplicate and the Kd was determined to be 0.075 nM (range 0.069-0.081 nM) (see FIG. 1).
[0099]Apilimod was also screened against a comprehensive panel of kinases (PIKfyve not included). In total, 456 kinases, including disease-relevant kinases, were assayed for their ability to bind with apilimod. The screening concentration of apilimod was 1 mM, a concentration that is >10,000 times greater than the Kd for apilimod against PIKfyve. The results from the screen showed that apilimod did not bind to any of the 456 kinases tested.
[0100]Together, these results demonstrate that apilimod binds with high selectivity to a single cellular kinase, PIKfyve....
example 2
Induces Vacuolization and Disrupts Intracellular Traficking in Cells
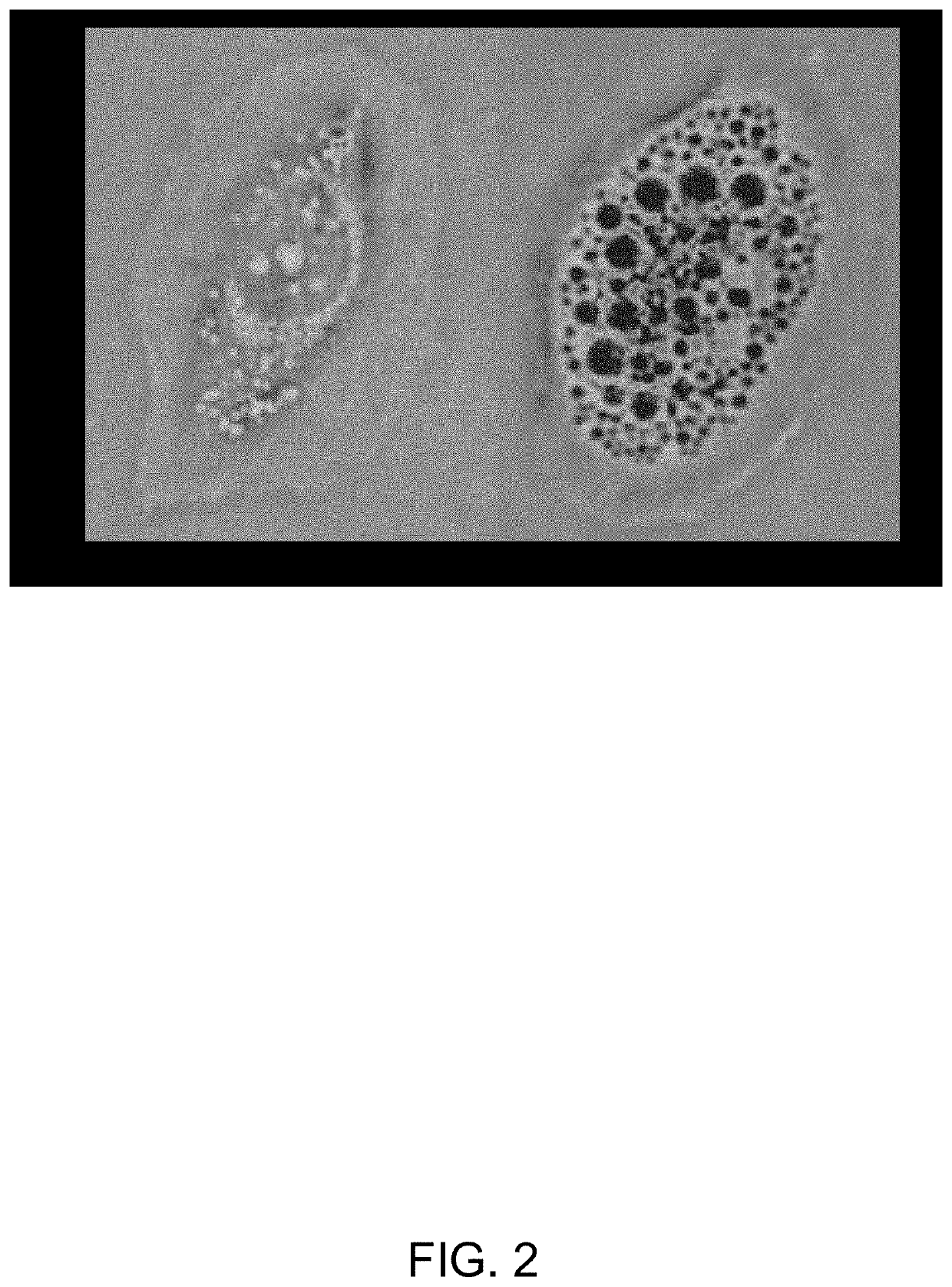
[0101]Apilimod has been demonstrated to be a potent and specific inhibitor of the phosphoinositide kinase PIKfyve, an enzyme that binds to PI(3)P and catalyzes the formation of the lipid second messengers PI(3,5)P2 and PI(5)P. PIKfyve is associated with the cytosolic leaflet of early endosomes and its activity is required for endomembrane homeostasis, endolysosomal function and proper retrograde transport from the endosome to the trans-Golgi network. Introduction of a kinase dead mutant into cells induces a swollen vacuole phenotype that can be rescued by the injection of PI(3,5)P2. Inhibition of PIKfyve by pharmacological methods as well as RNAi also produces swollen vacuoles and disruption of endomembrane dynamics. It has been discovered that pharmacological disruption of PIKfyve with apilimod induces selective lethality of specific cancer cell lines through disruption of intracellular trafficking (see FIG. 2).
example 3
n of in vivo Anti-Ebola Activity in Humans
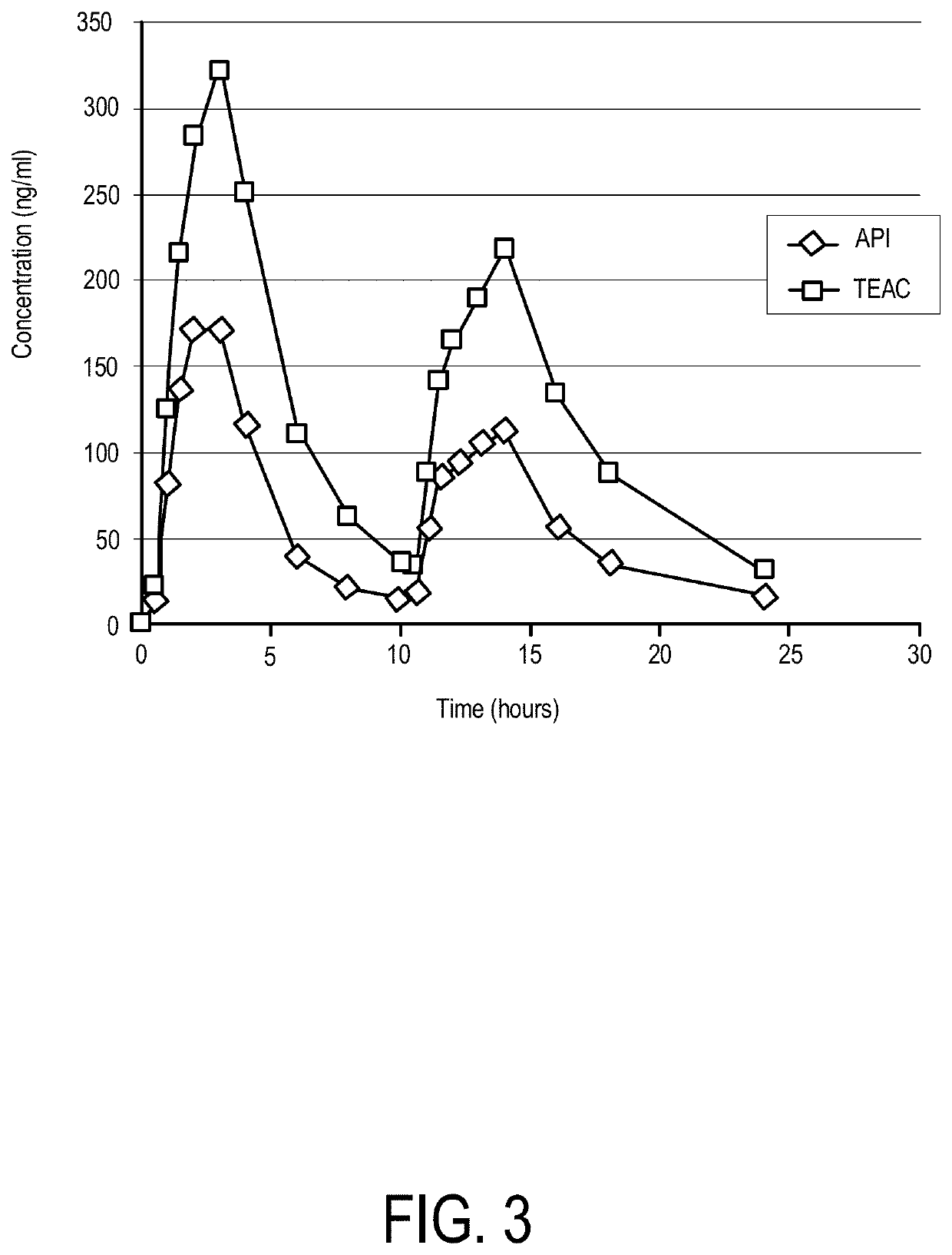
[0102]Inhibition of cancer cell proliferation and inhibition of Ebola virus infection share a common mechanism i.e. inhibition of PIKfyve leading to vacuole formation and loss of intracellular trafficking. In the above clinical study, the trough TAEC values were greater than 25 mg / mL (60 nM). Since vacuole formation in cells at apilimod concentrations as low as 20 nM have been observed, one could conclude that oral administration of apilimod free base at doses ranging from 70 to 1000 mg / day should provide continuous PIKfyve inhibition to maintain vacuolization of cells and block Ebola infection in clinical therapy in patients.
[0103]Furthermore, it was shown in female Balb / c mice, a strain often used for in vivo Ebola infection studies, that constant infusion of apilimod as the bis-mesylate salt, using subcutaneously implanted osmotic mini-pumps (e.g. Alzet models 1007D, 15 mg / kg / day; model 2001, 30 mg / kg / day; vehicle: 25% DMSO, 25% Cremaphor...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com