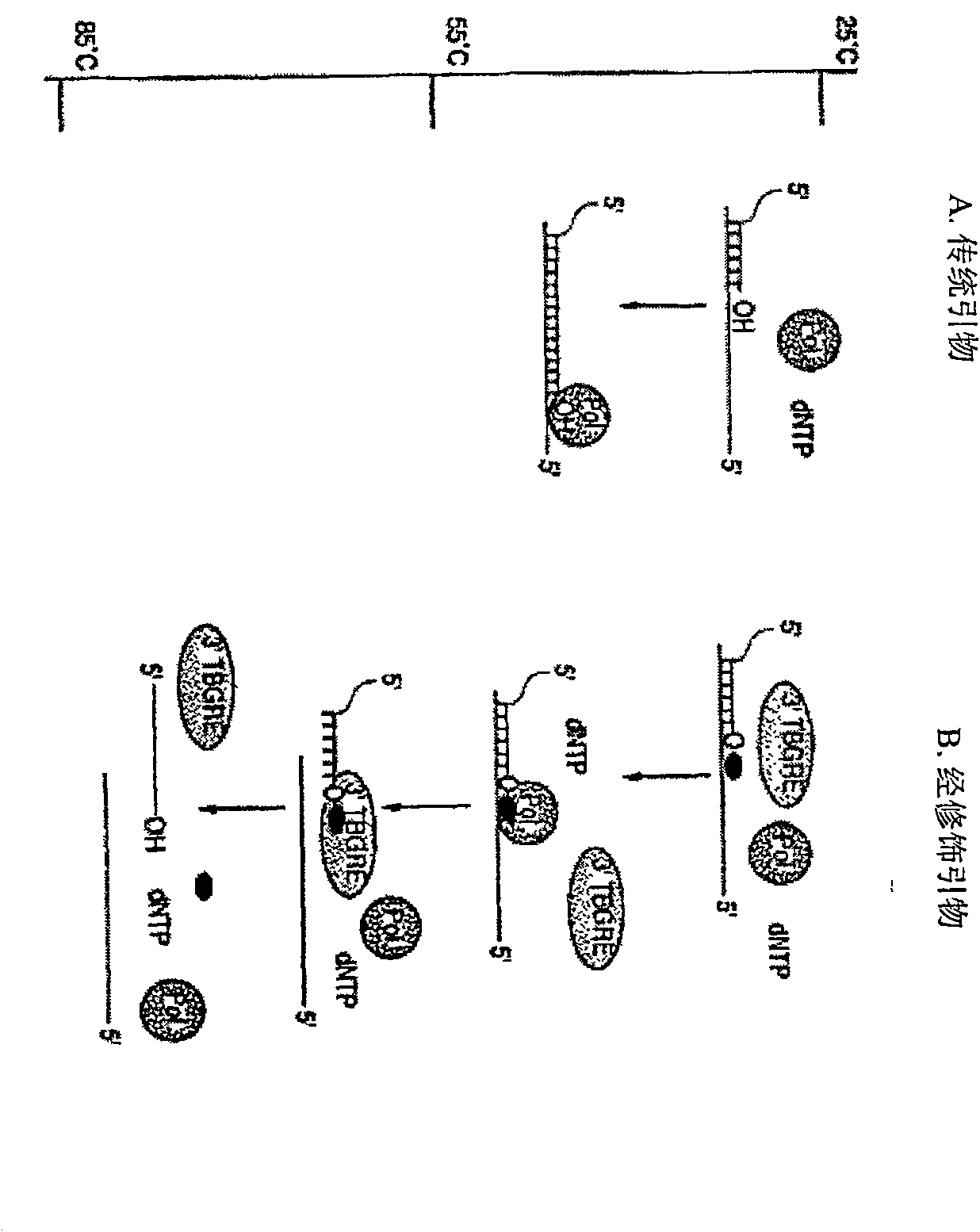
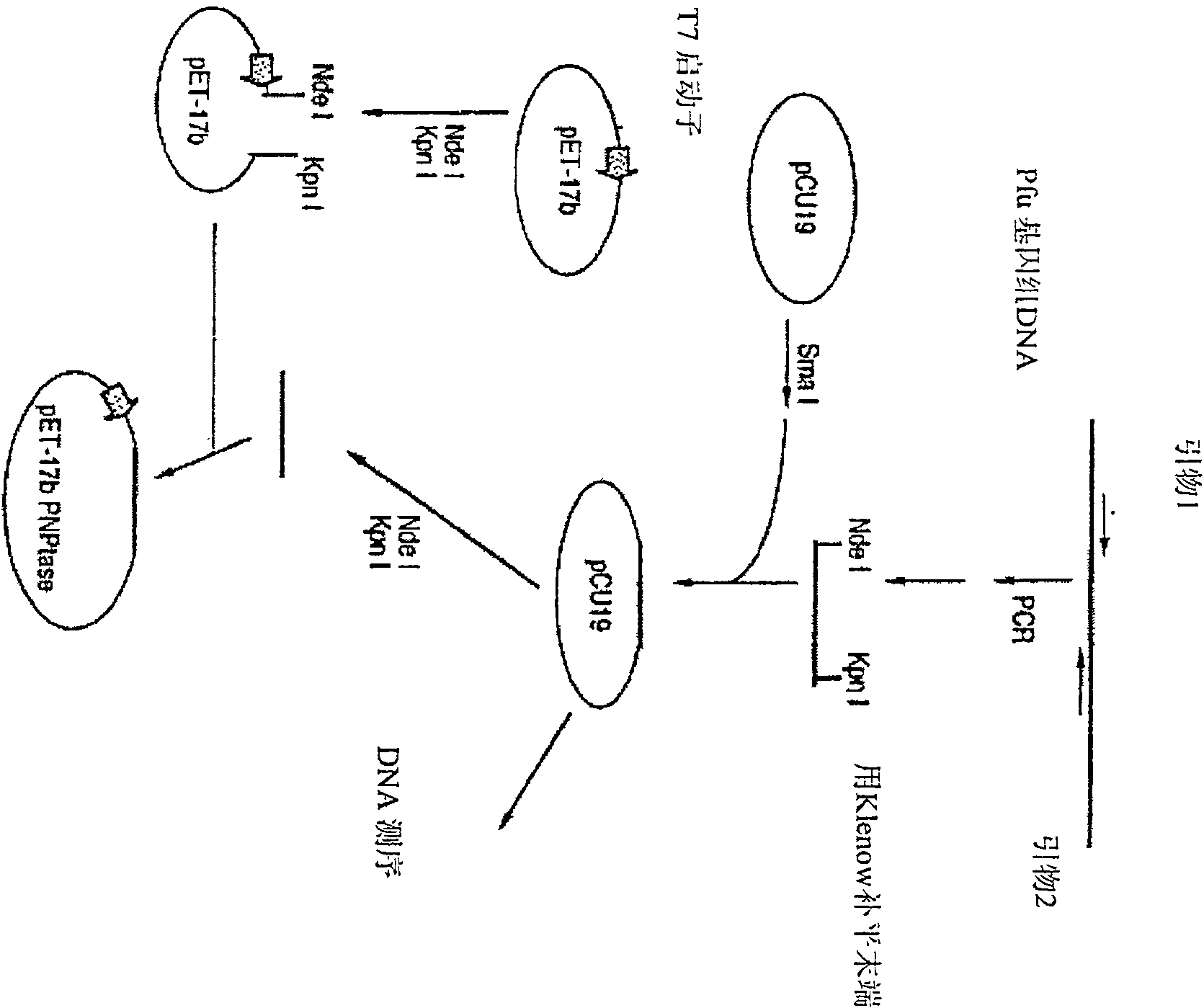
Method and compositions for improved polynucleotide synthesis
A polynucleotide and nucleoside technology, applied in microorganism-based methods, plant genetic improvement, chemical instruments and methods, etc., can solve problems such as enzyme temperature sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0102] 3′polynucleotide phosphatase activity detection
[0103] 3'phosphate-modified oligonucleotides were chemically synthesized (Midland Certified Reaget Company, Midland, Tex.). The oligonucleotide sequence consists of the following:
[0104] 5'-GCT GCTCTGTGCATCCGAGTGG-p-3' (SEQ ID No: 7)
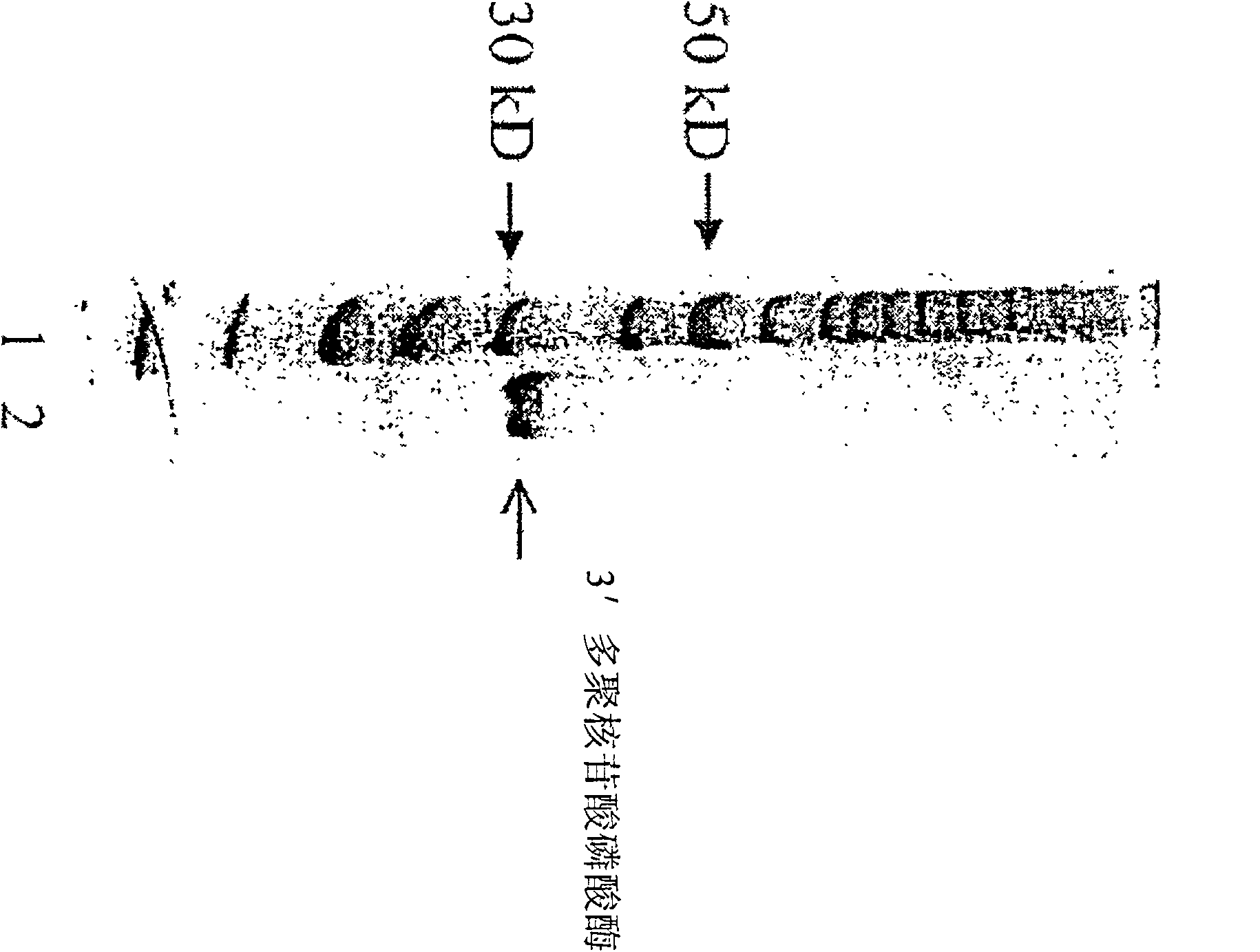
[0105] 32P was labeled at the 5' end of the oligonucleotide with T4 polynucleotide kinase which has no 3' polynucleotide phosphatase activity (Yang, S.W., et al.Proc.Natl.Acad.Sci.U.S.A.93(21) : 11534-9 (1996)). with 32 P-labeled oligonucleotides were mixed with purified 3′ polynucleotide phosphatase in 1×PCR reaction buffer (10Mm Tris-HCl, pH8.3, 50mM MgCl 2) at 72°C for 5 minutes. Add DNA sequencing buffer, heat at 90° C. for 2 minutes, and analyze the sample by 12% polyacrylamide-7M urea gel electrophoresis. Figure 4 is an autoradiogram of samples quantified with PhophorImager (Molecular Dynamics). One unit of 3' phosphatase is defined as the ability to remove 5 μmol of the 3' ...
example 2
[0107] Isolation and Purification of 3' Phosphatase from Pfu
[0108] 100 g wet weight of Pfu cells (purchased from the Marine Biotechnology Center at the University of Maryland, Baltimore, Md) were suspended in lysate, 20 mM Tris-HCl, Ph7.5, 1 mM EDTA, 1 mM DTT, 0.2 M NaCl on ice , 10 mM mercaptoethanol and 2 mM benzylsulfur fluoride. Cells were then lysed by ultrasonication and centrifuged at 8,200 rpm for 10 minutes on a Sorvall GS-3 rotor. Add 0.05 vol of 10% polyethyleneimine solution to the supernatant, mix well and centrifuge, then fractionate with ammonium sulfate (45-80% saturation). Then use phosphocellulose column (P-11; Whatman, Inc.; activity eluted.apprxeq.0.6M NaCl), Source 15S (Pharmacia; activity eluted.apprxeq.0.2M NaCl), double-stranded DNA-cellulose (Sigma; activityeluted.apprxeq.0.15M NaCl), Mono S column (Pharmacia; activityeluted.apprxeq.0.35M NaCl) and heparin agarose, Mono Q column (Pharmacia; activityeluted.apprxeq.0.25M NaCl) chromatography, finall...
example 3
[0111] Peptide sequencing of the N-terminus of 3′ phosphatases
[0112] Part of the purified 3' polynucleotide phosphatase was added to 7% SDS-Tricine PAGE. The protein in the SDS gel was electrotransferred to PVDF membrane (Immobolin-P from Millipore) in transfer buffer (0.5.times.TBE (pH 8.4), 20% methanol, 0.5mMEDTA), 0.5A electrophoresis, transfer 1 Hour. Membranes were stained with Coomassie Brilliant Blue R-250 for 10 minutes and destained twice with 100% methanol. The band corresponding to the 3' polynucleotide phosphatase on the membrane was excised. Its amino acid sequencing was done with Yale University commercial facilities. List the sequence of 26 amino acids in single letters starting from the N-terminus:
[0113] FKIDRLRFGTAGIPLSTPKPSTIAGI (SEQ ID NO: 1).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com