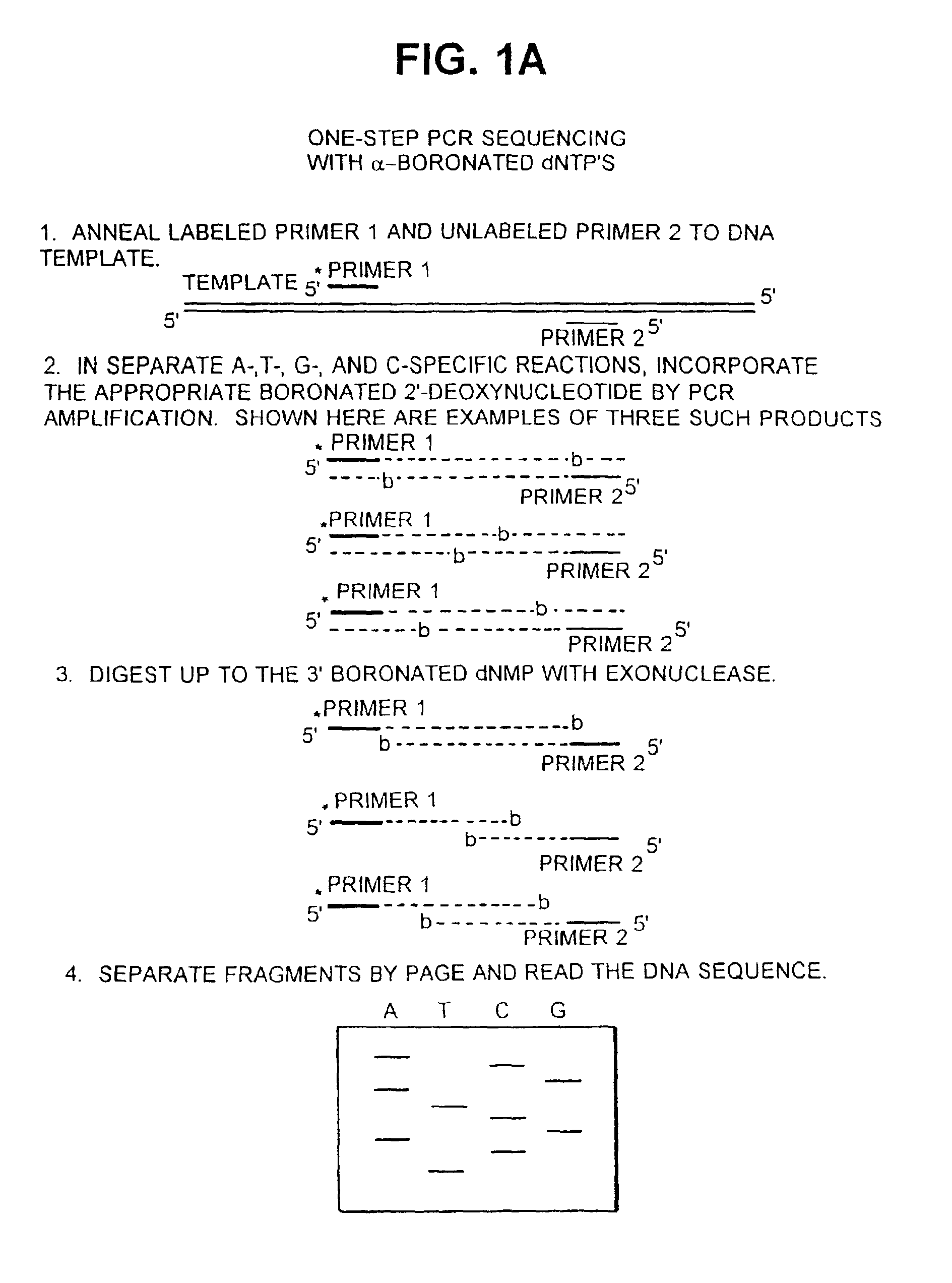
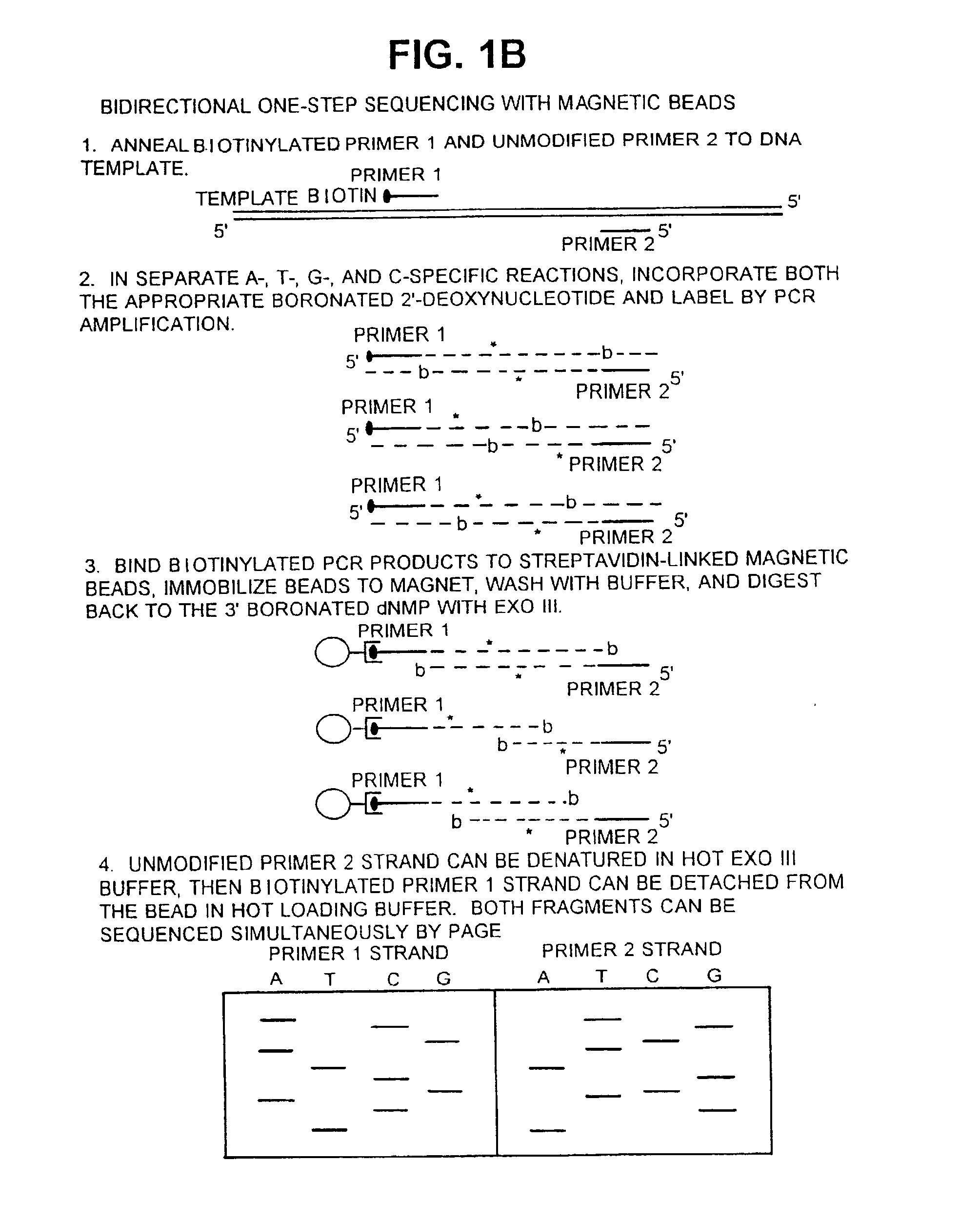
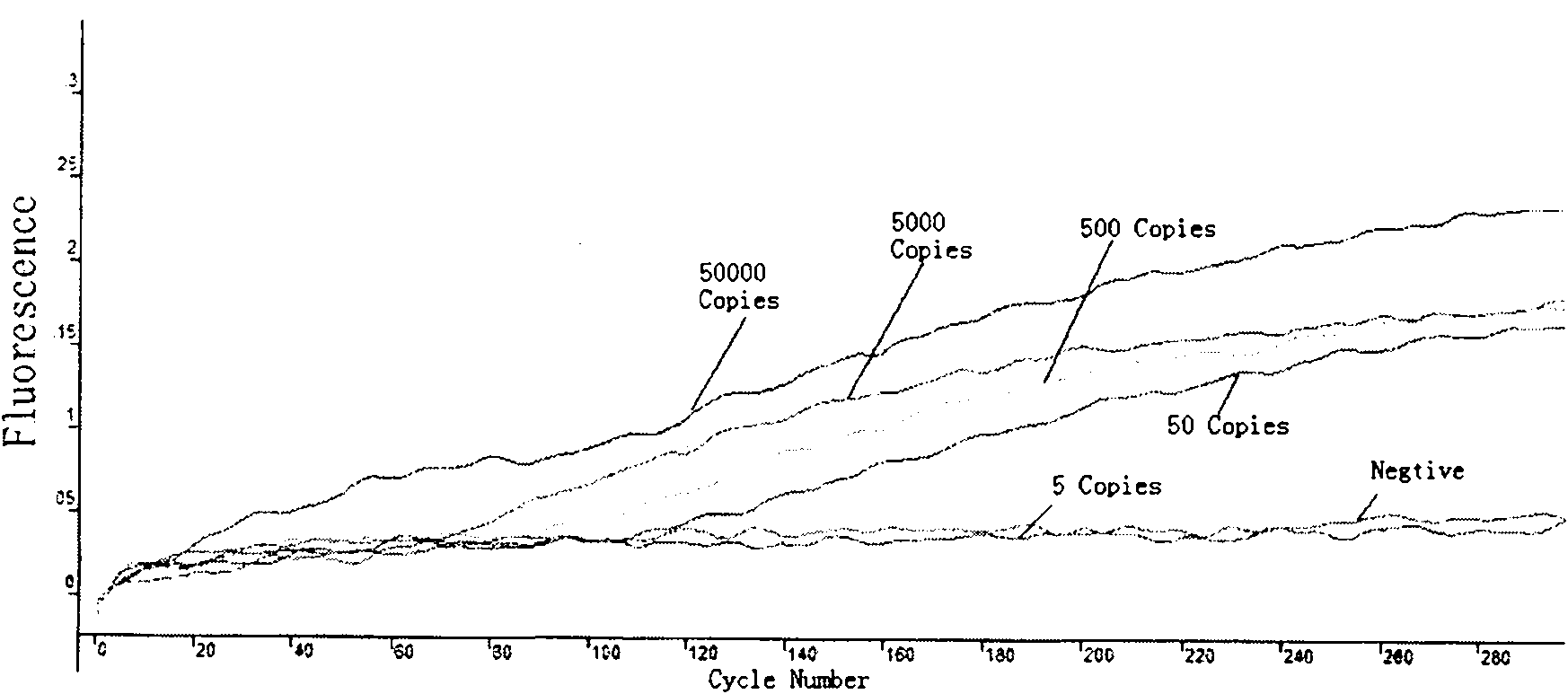
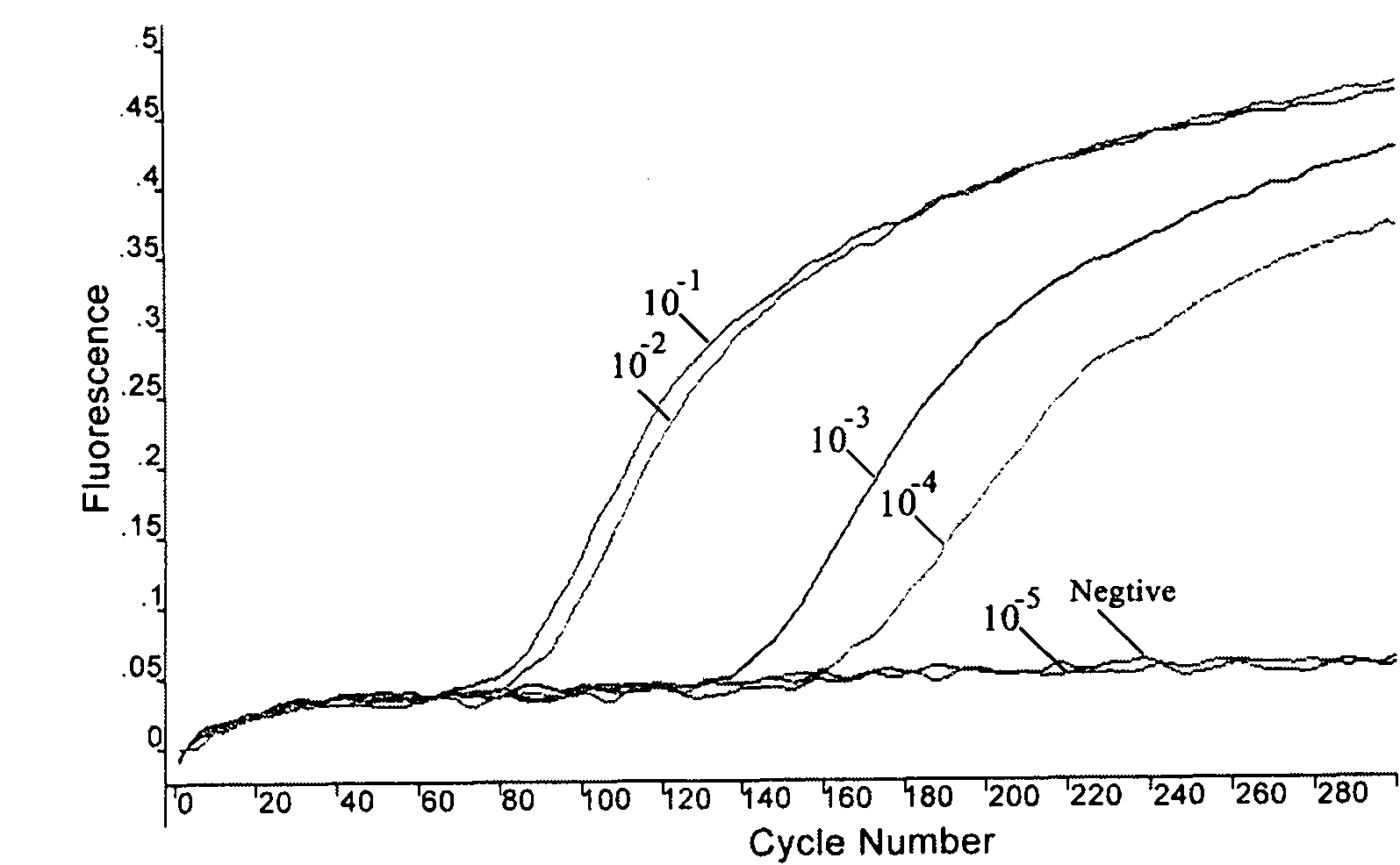
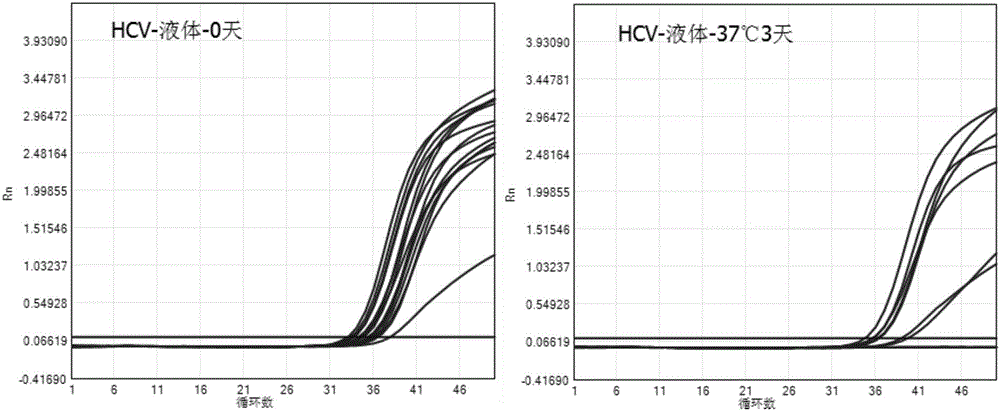
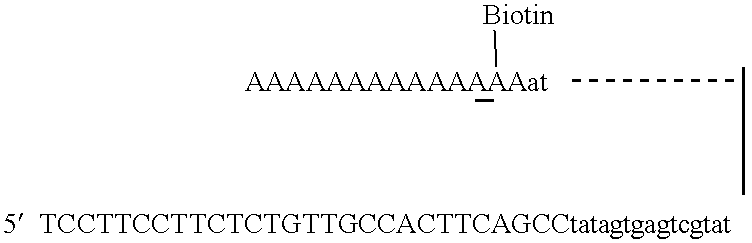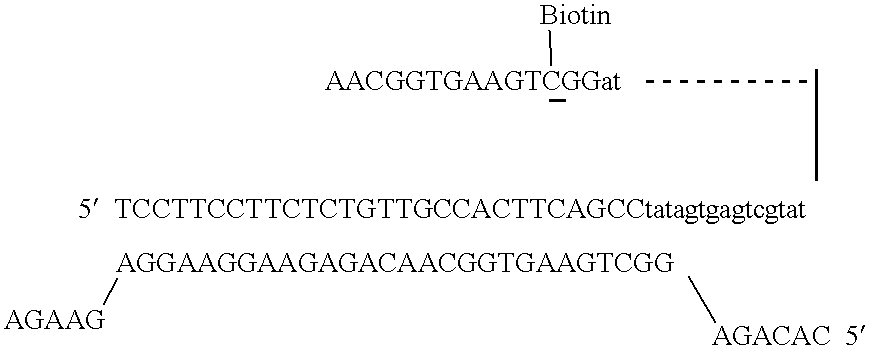Patents
Literature
95 results about "RNA amplification" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
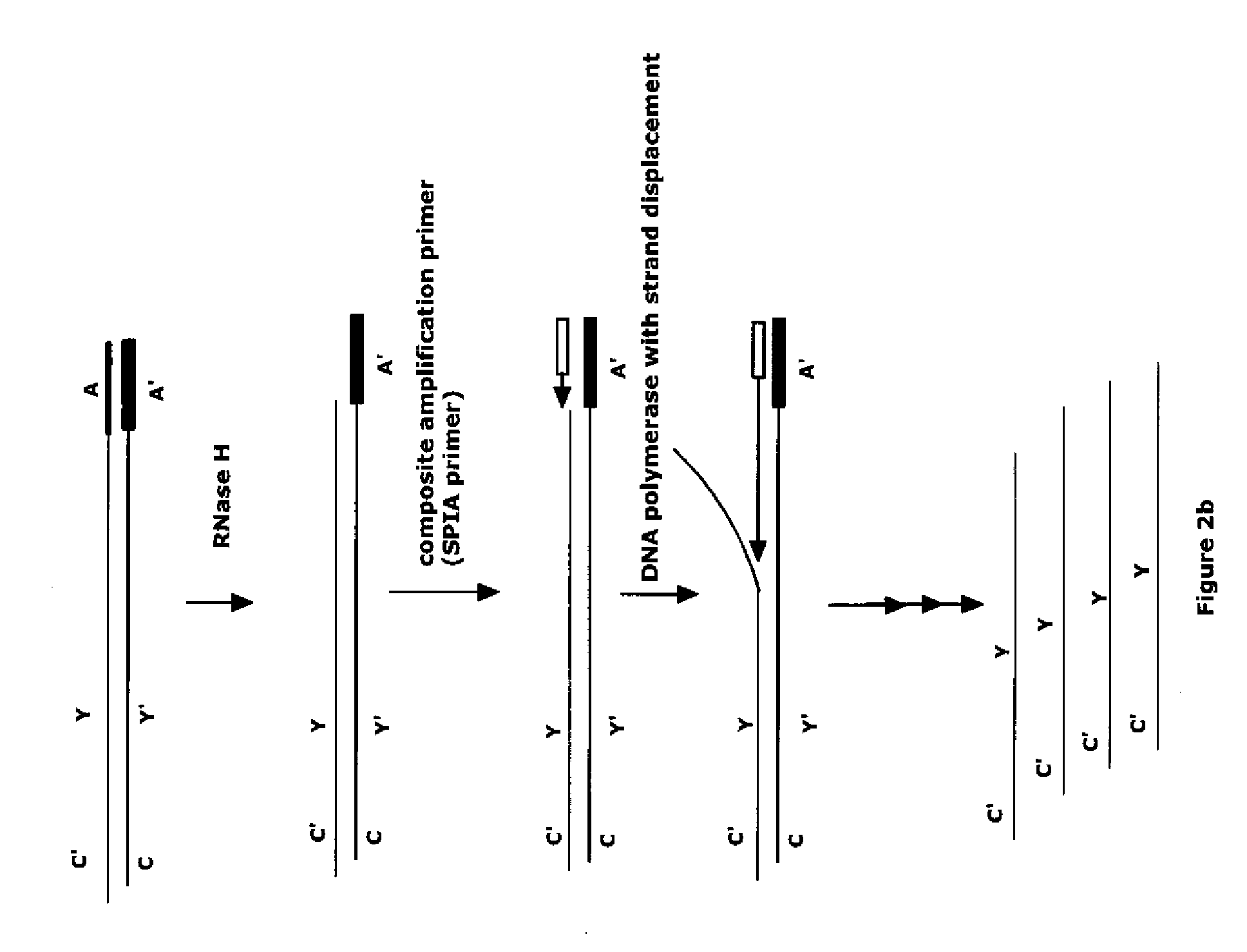
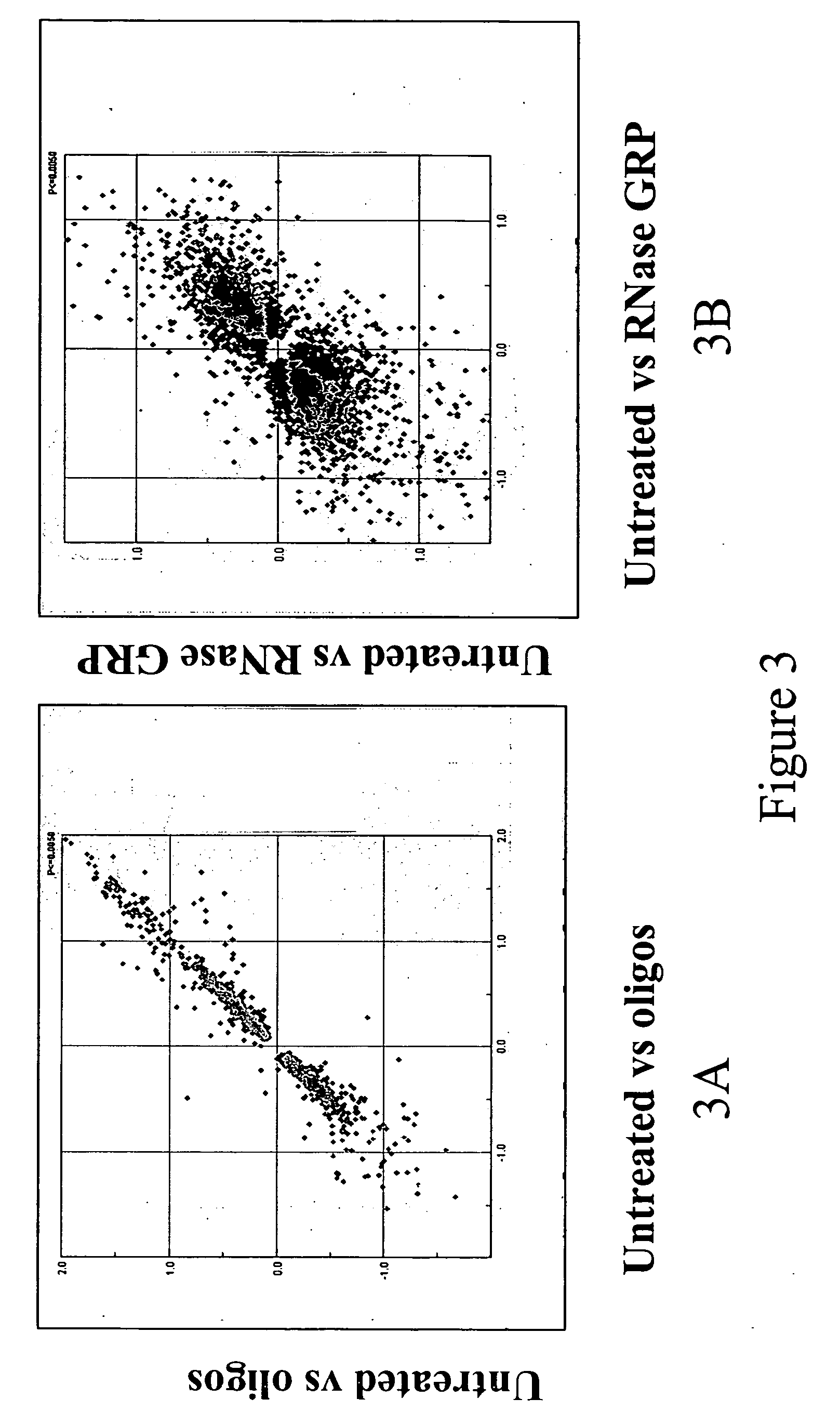
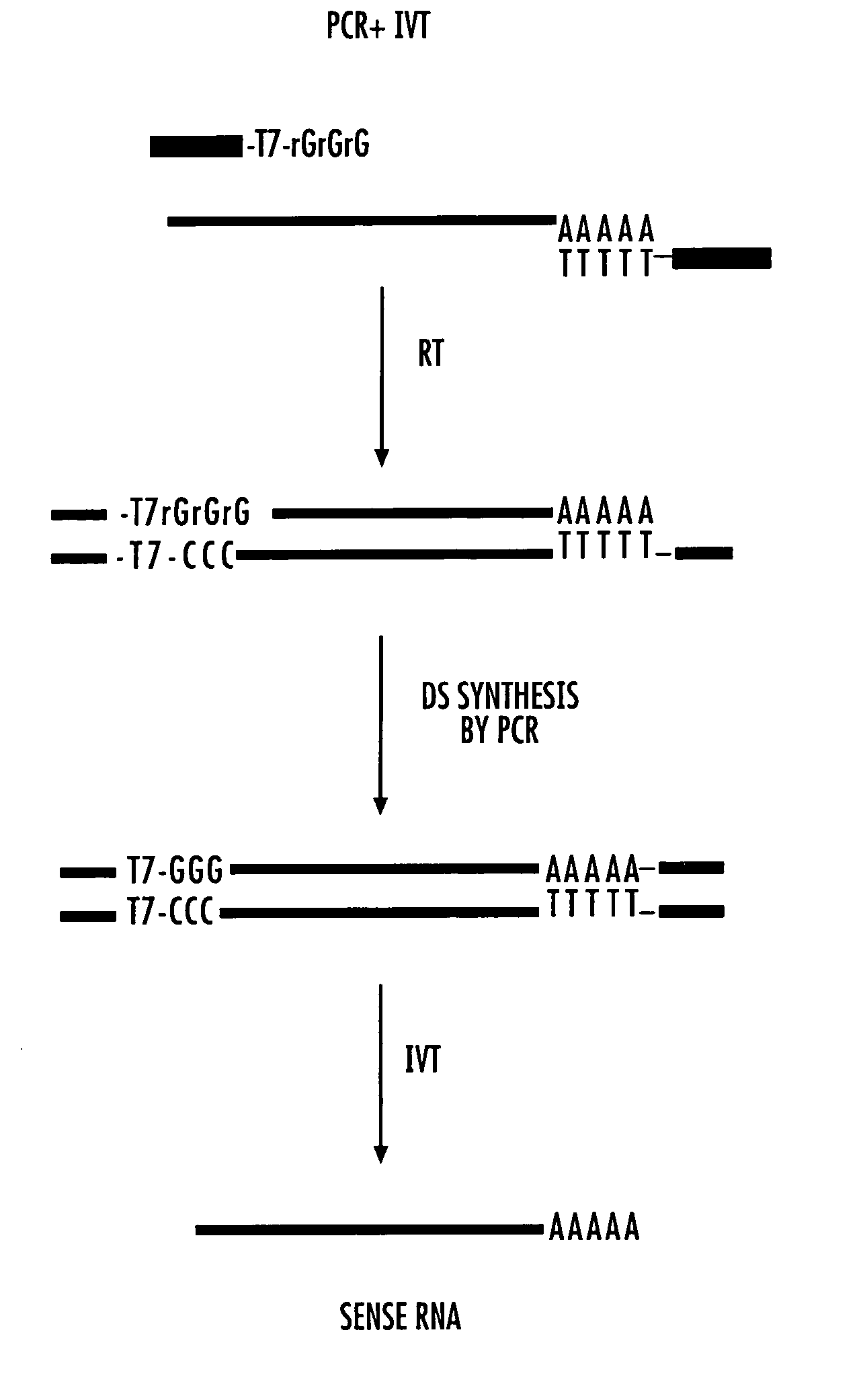
With the SeqPlex RNA amplification kit, the RNA samples were reverse transcribed with random primers having semidegenerate 3′ ends and defined universal 5′ ends (Supplemental Fig. 4). The displaced single strands generated during the process serve as a template for primer annealing and second-strand cDNA synthesis.
Recombinase polymerase amplification
ActiveUS8071308B2Lower Level RequirementsImprove abilitiesSugar derivativesMicrobiological testing/measurementLoading factorRecombinase Polymerase Amplification
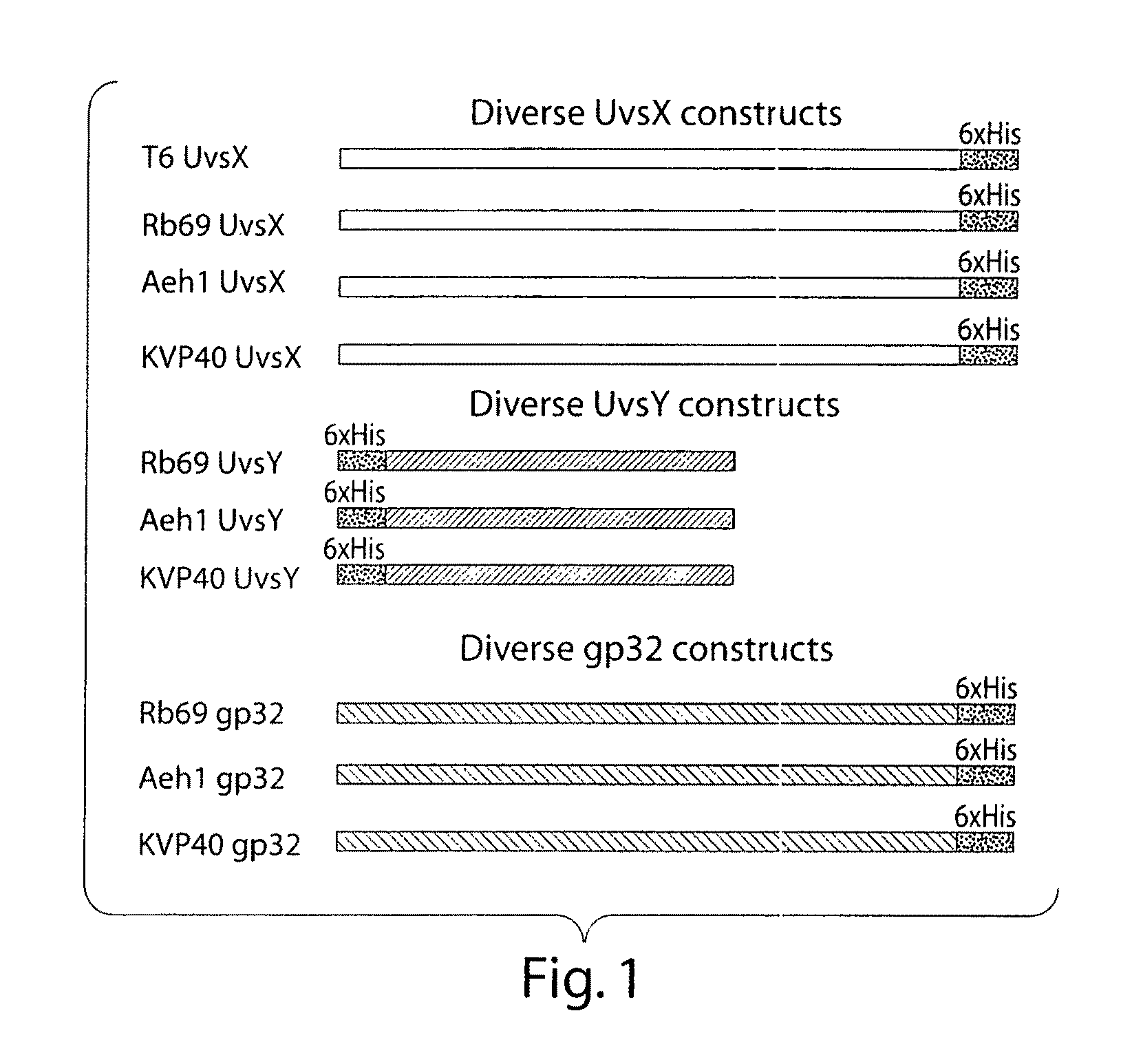
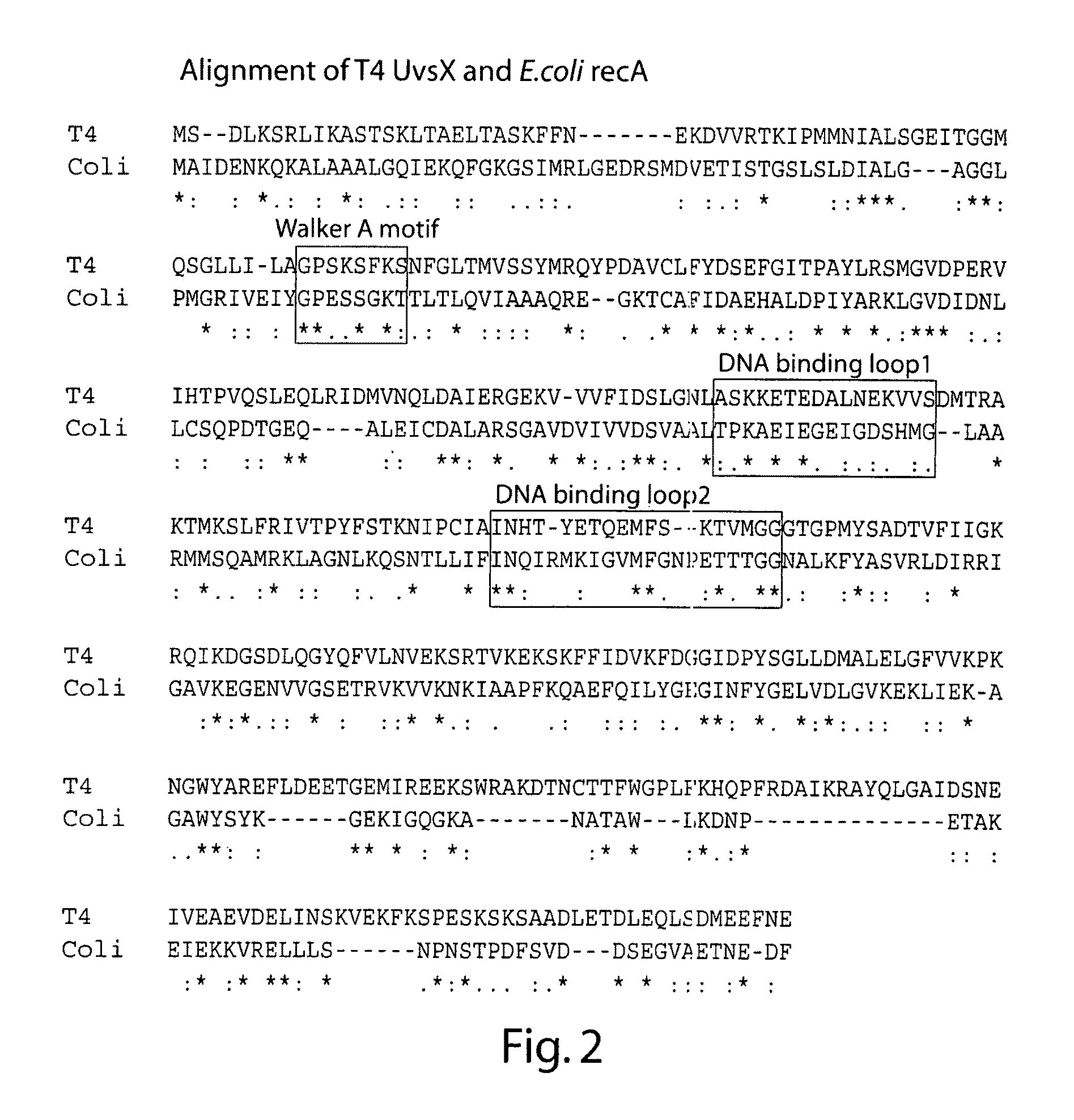
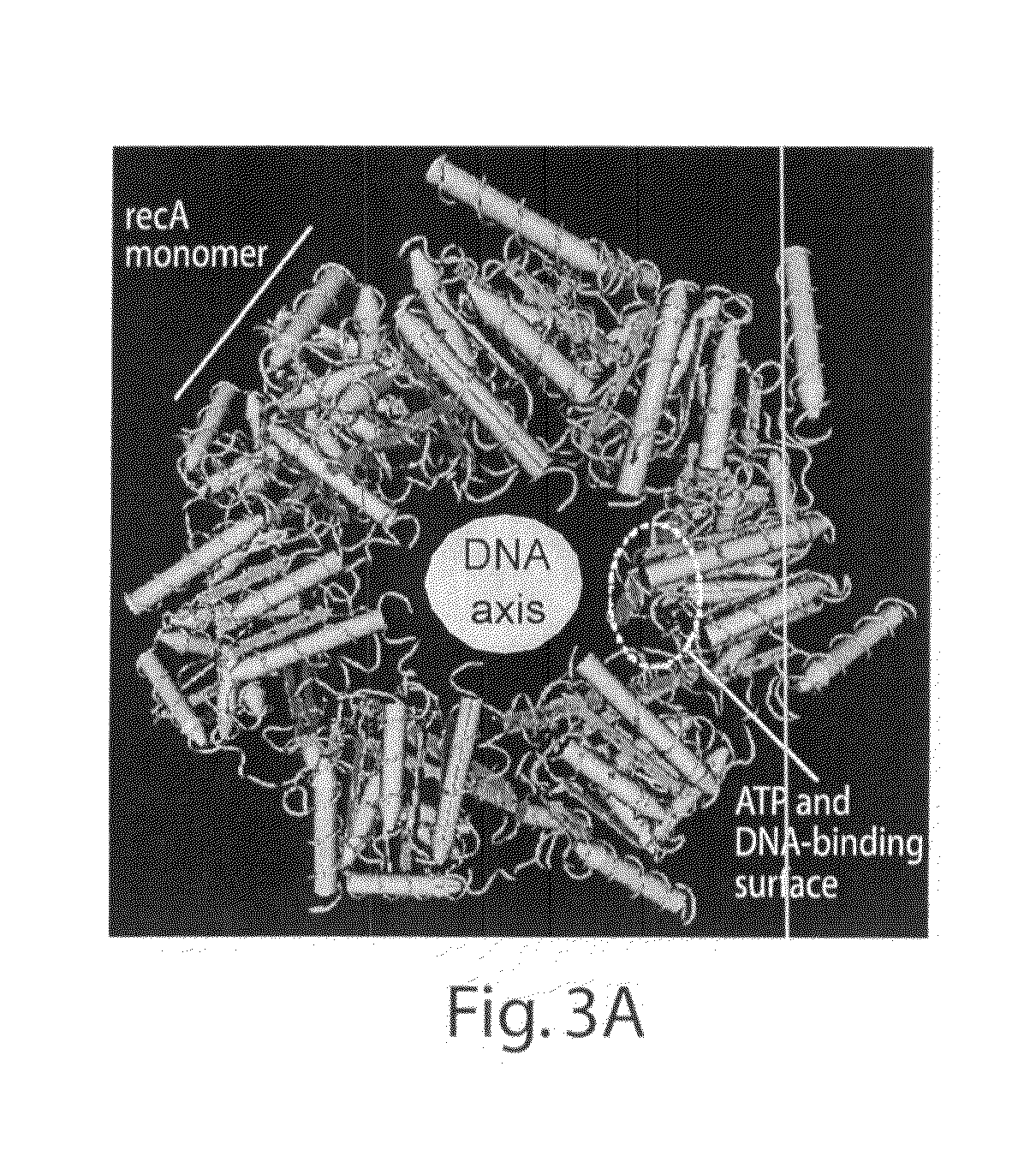
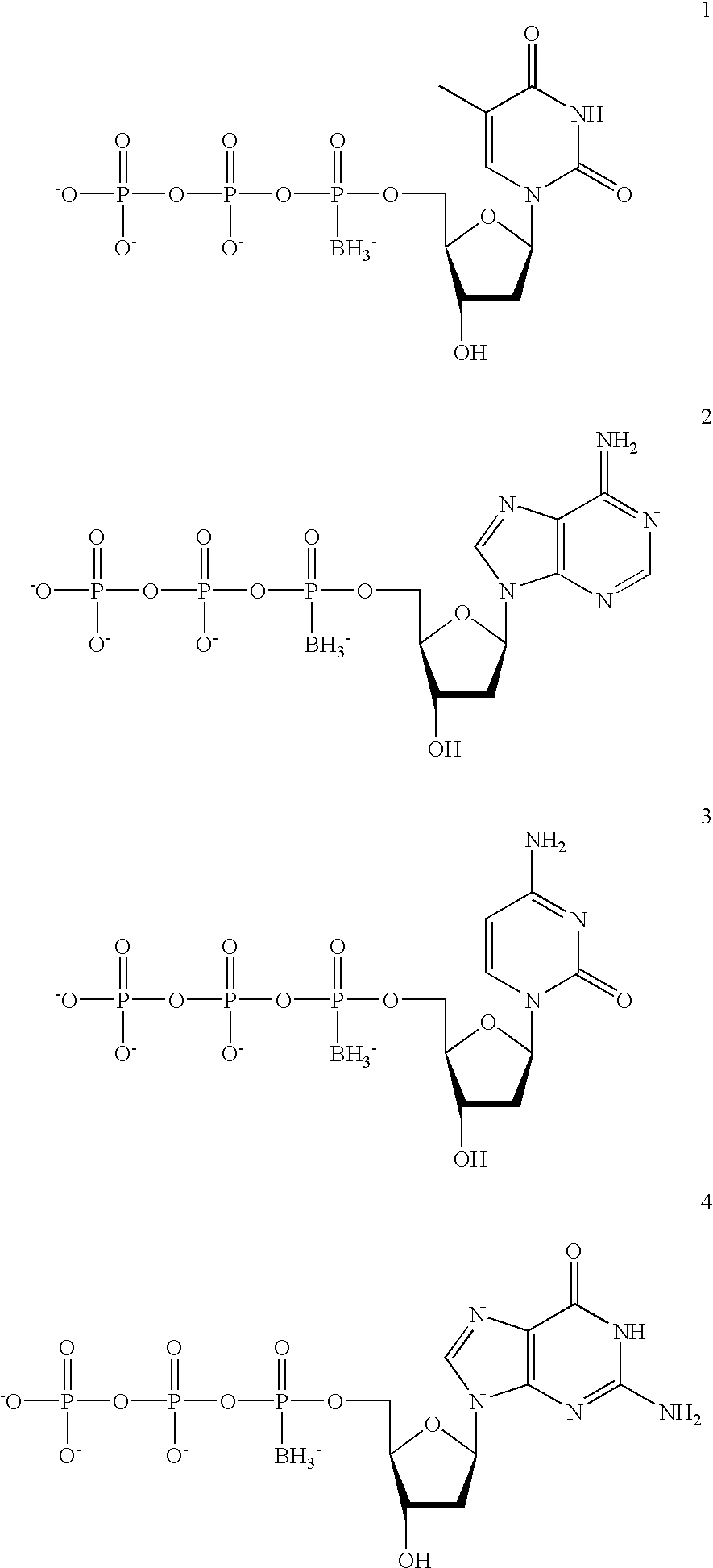
The present invention features novel, diverse, hybrid and engineered recombinase enzymes, and the utility of such proteins with associated recombination factors for carrying out DNA amplification assays. The present invention also features different recombinase ‘systems’ having distinct biochemical activities in DNA amplification assays, and differing requirements for loading factors, single-stranded DNA binding proteins (SSBs), and the quantity of crowding agent employed.
Owner:ABBOTT DIAGNOSTICS SCARBOROUGH INC
Method of nucleic acid sequencing
InactiveUS20030064483A1High yieldSugar derivativesPeptide/protein ingredientsNucleotideNucleic acid sequencing
The present invention relates, in general, to a process of enzymatically synthesizing nucleic acids containing nucleotides that are resistant to degradation. The invention further relates to methods of utilizing such nucleic acids in DNA and RNA amplification and sequencing, gene therapy and molecular detection protocols.
Owner:DUKE UNIV
Methods of RNA amplification in the presence of DNA
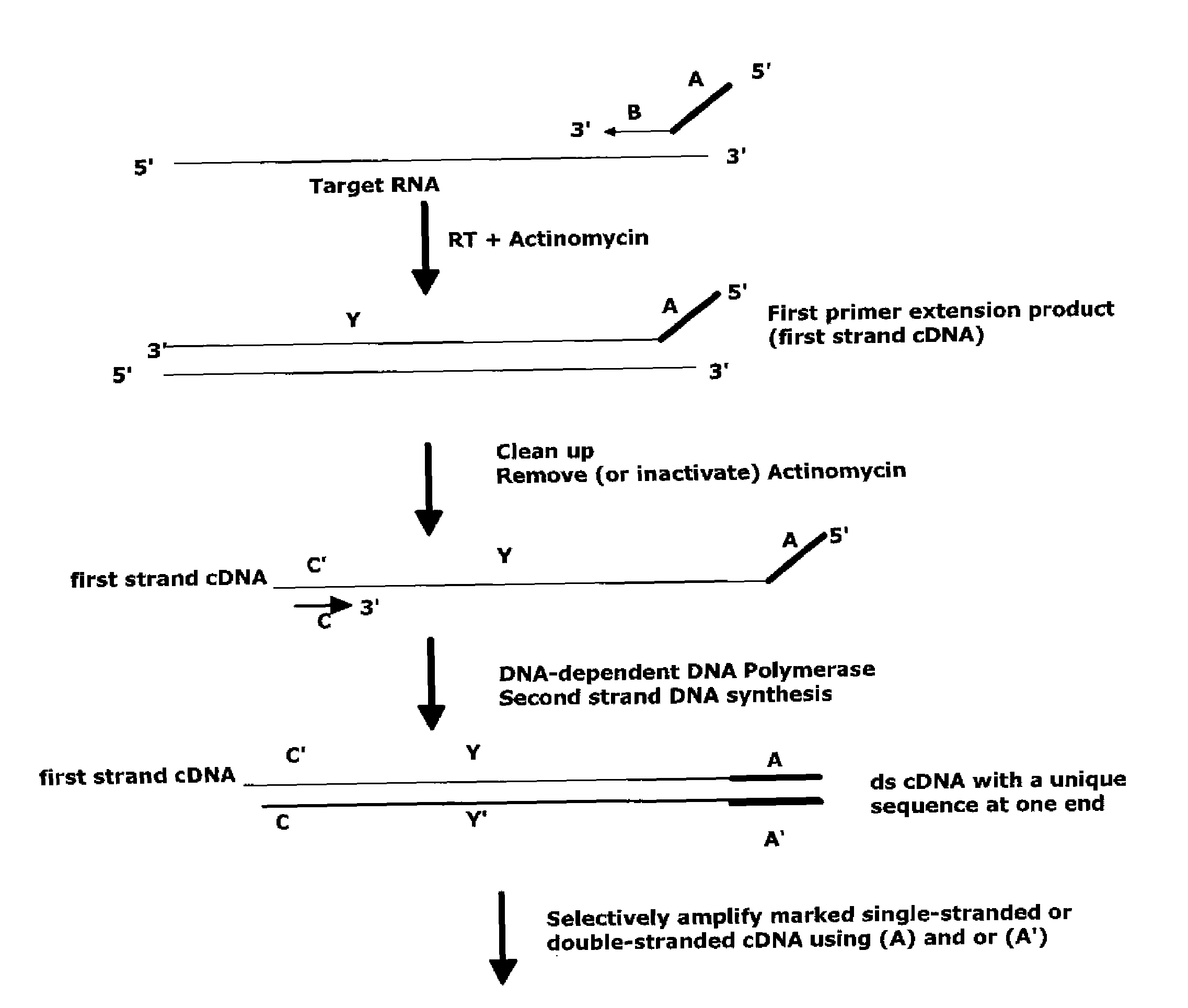
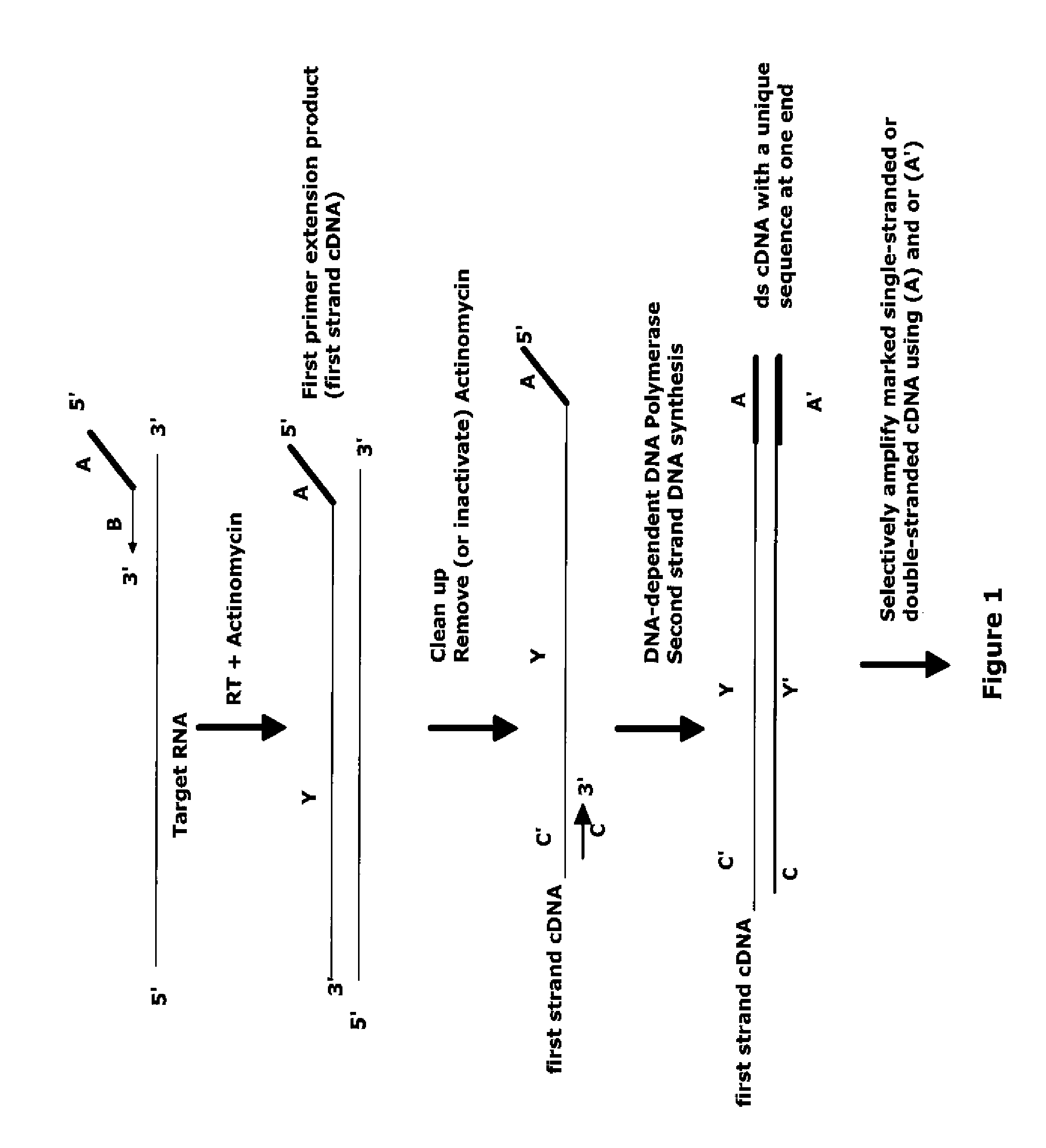
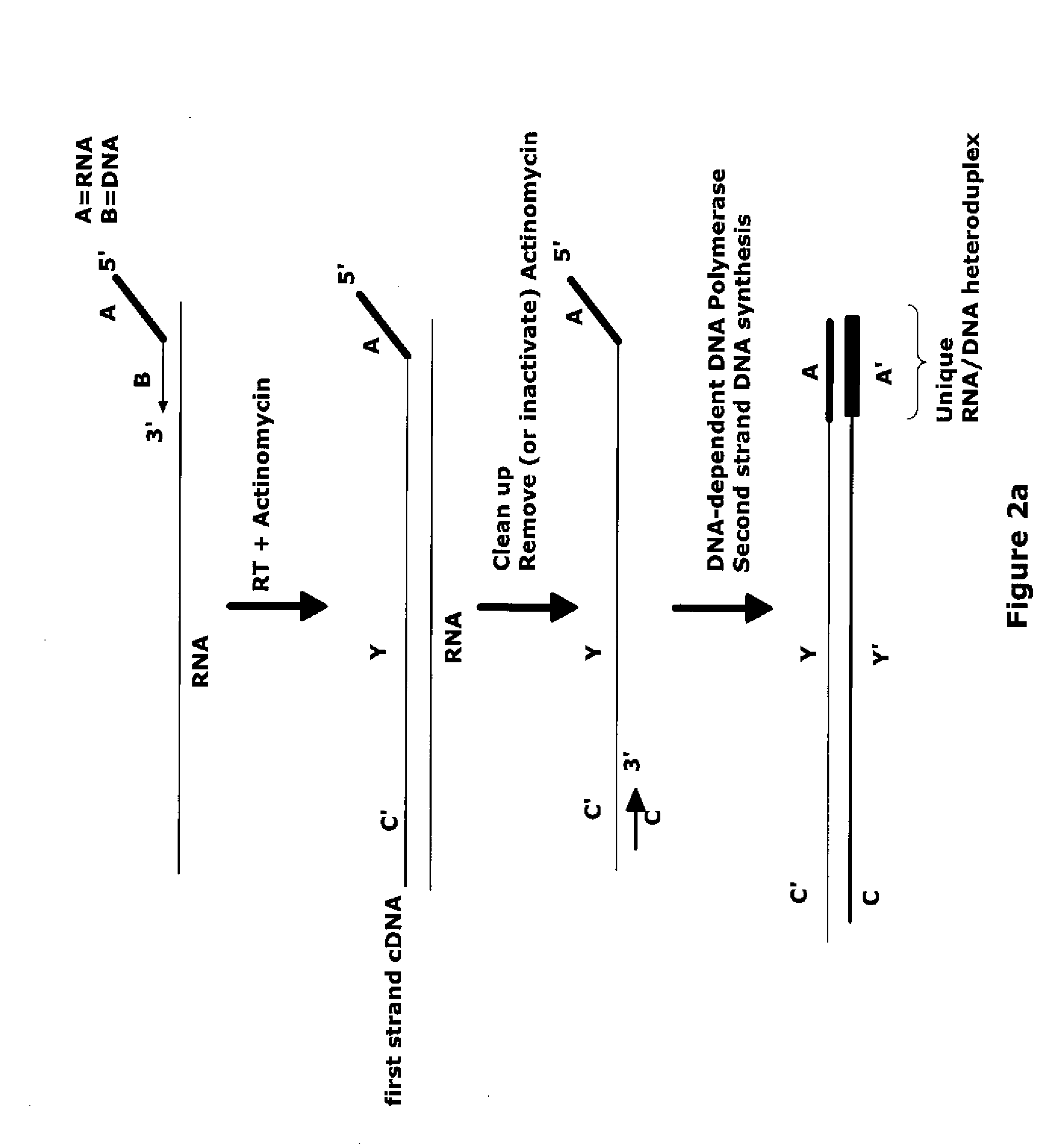
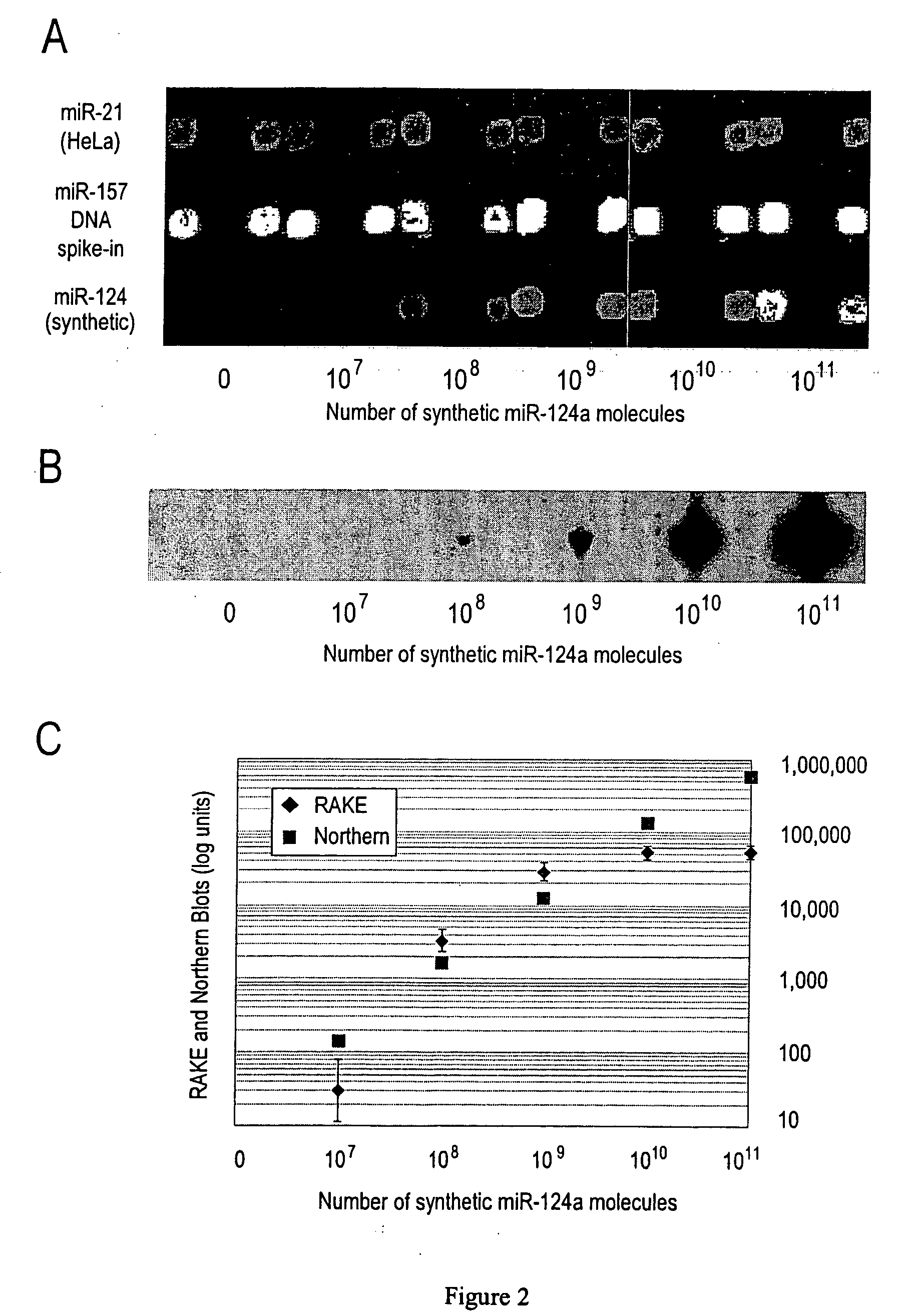
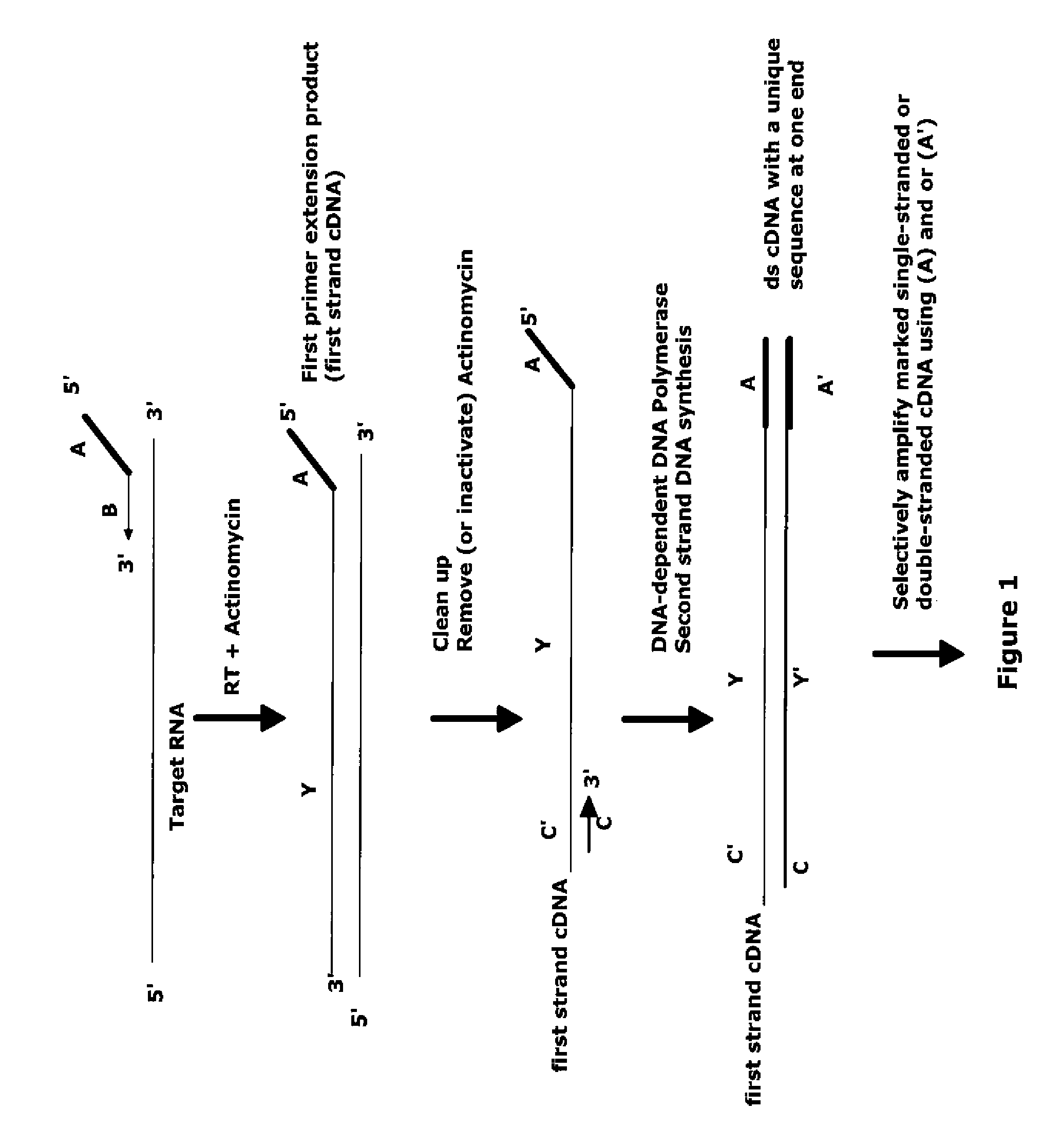
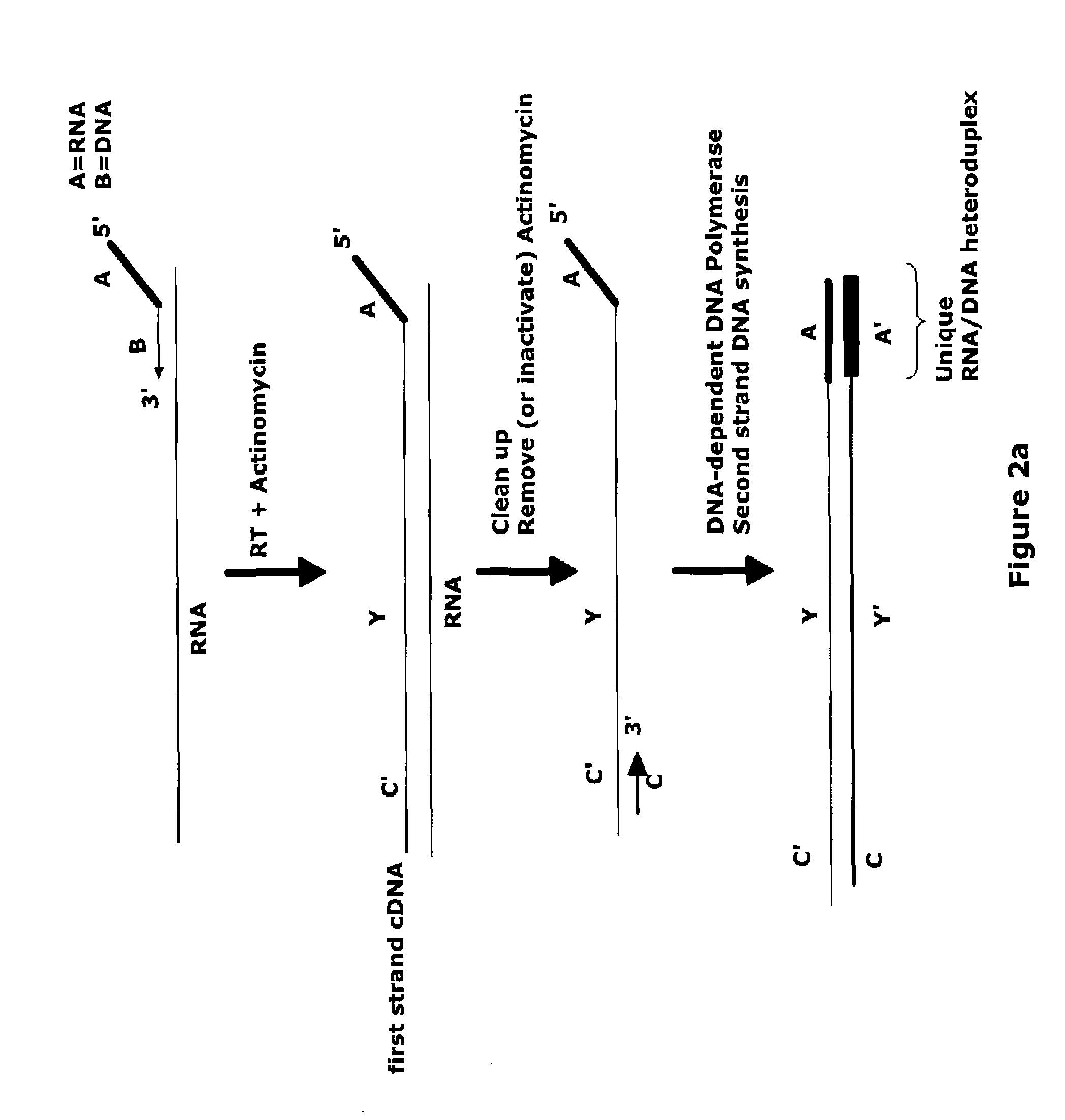
The invention provides methods for amplification of RNA. The methods are particularly suitable for specifically amplifying RNA in the presence of DNA. The methods involve producing a marked first primer extension product from a target RNA in the presence of a DNA-dependent DNA polymerase inhibitor, which prevents replication of DNA by the reverse transcriptase enzyme. The marked nucleic acid products are subsequently selectively amplified in the presence on non-marked nucleic acids. The methods are useful for production and analysis of polynucleotide sequences complementary to an RNA sequence. The methods are useful for preparation of nucleic acid libraries and substrates for analysis of gene expression of cells in biological samples. The invention also provides compositions and kits for practicing the amplification methods, as well as methods which use the amplification products.
Owner:NUGEN TECH
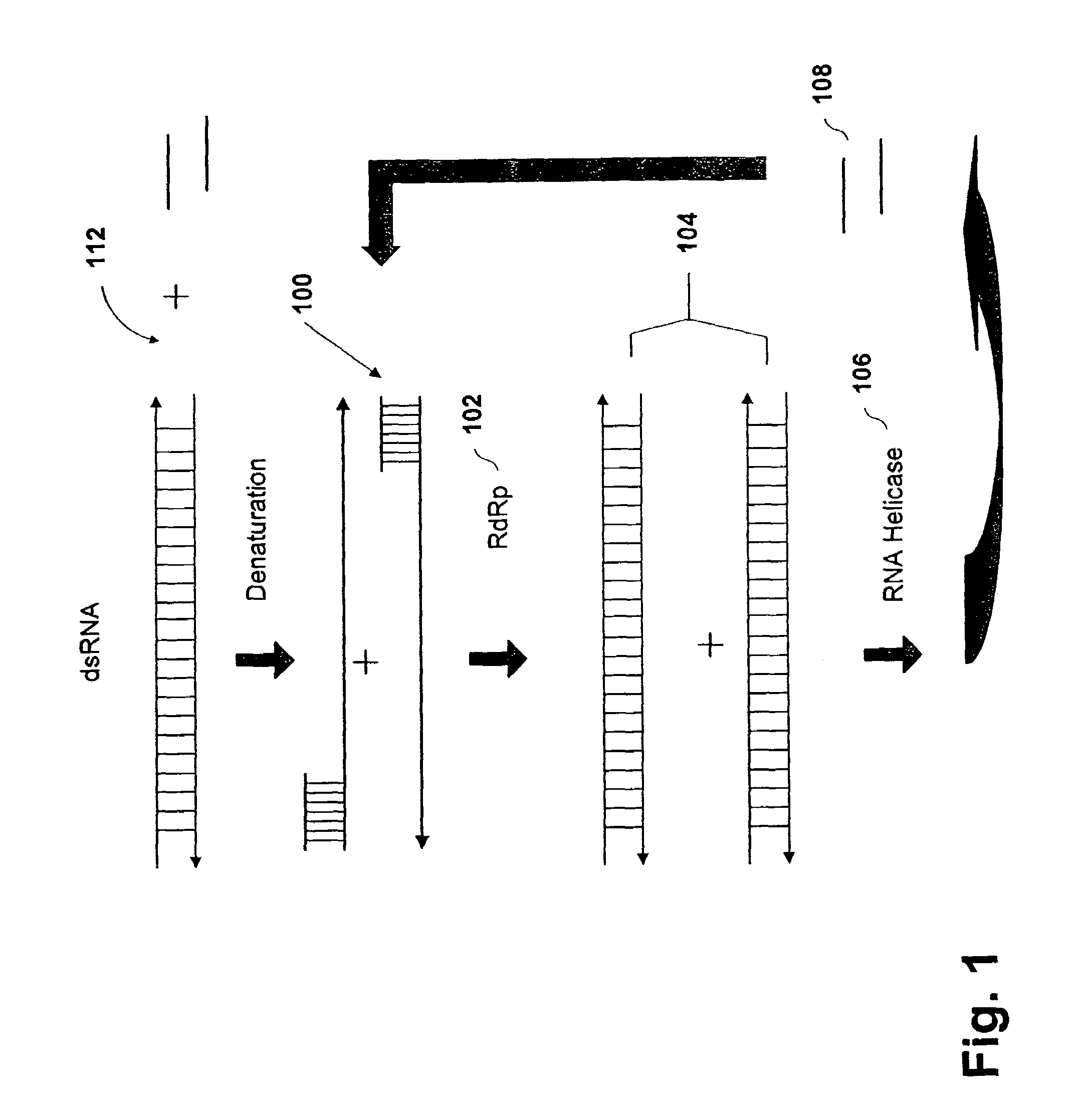
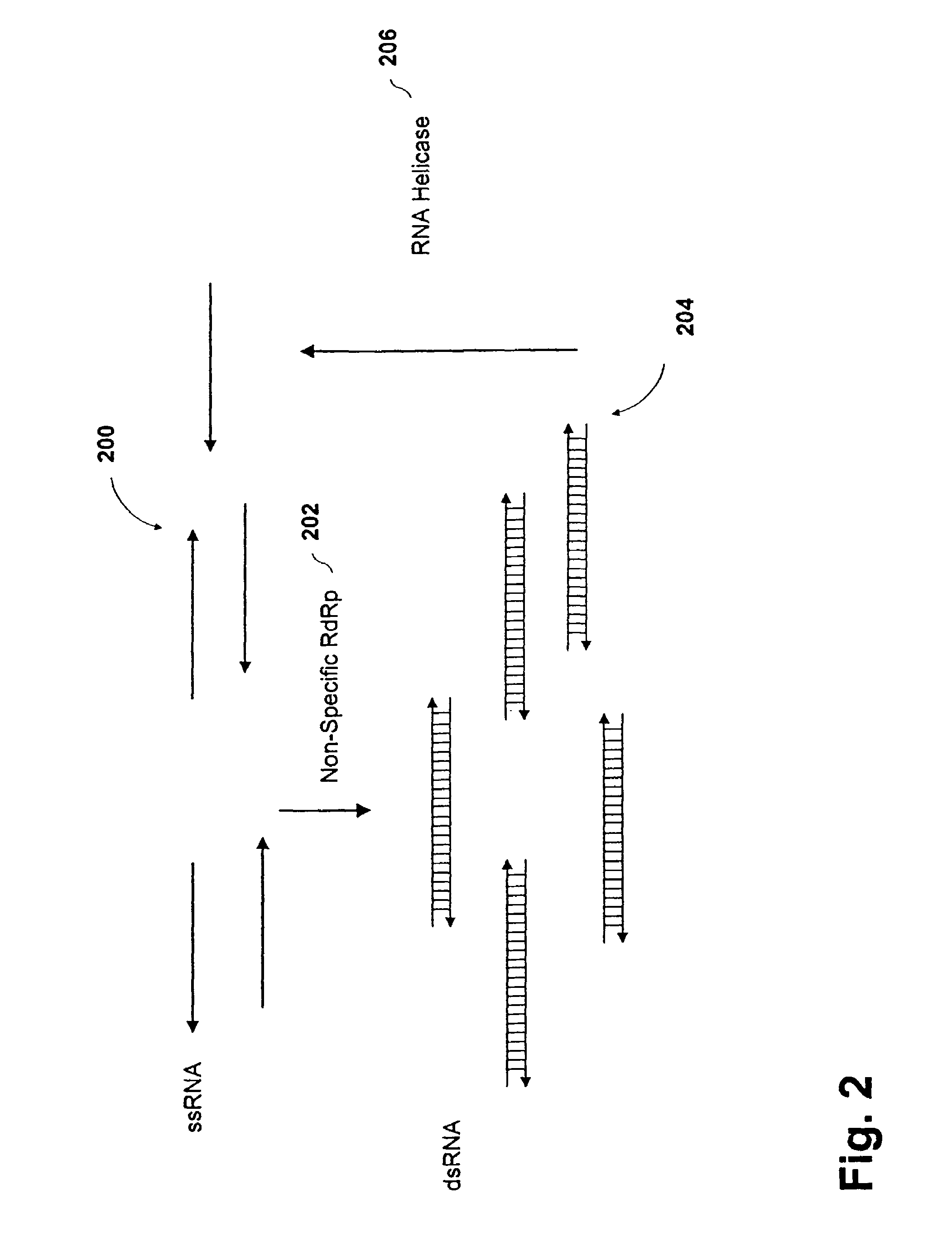
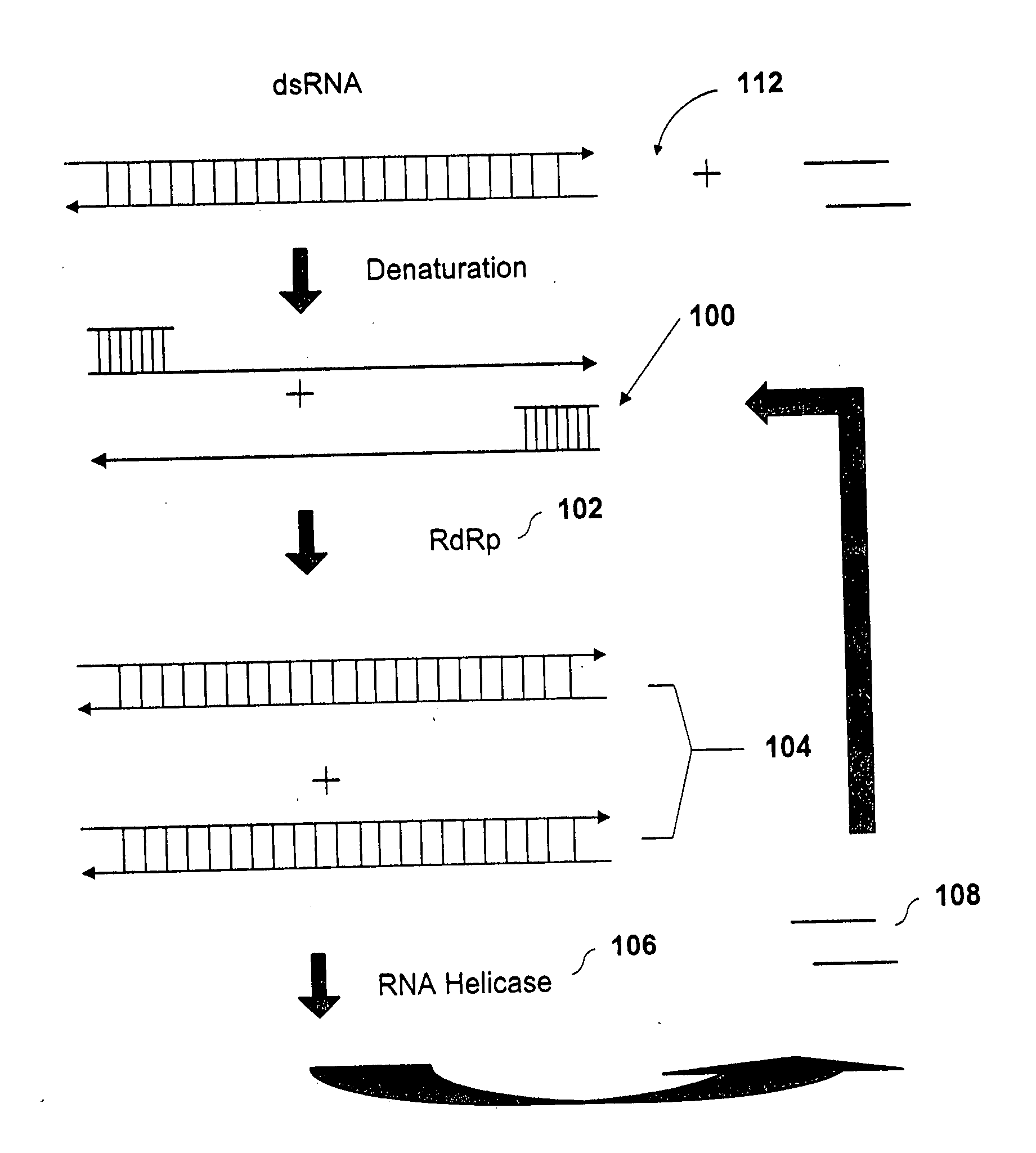
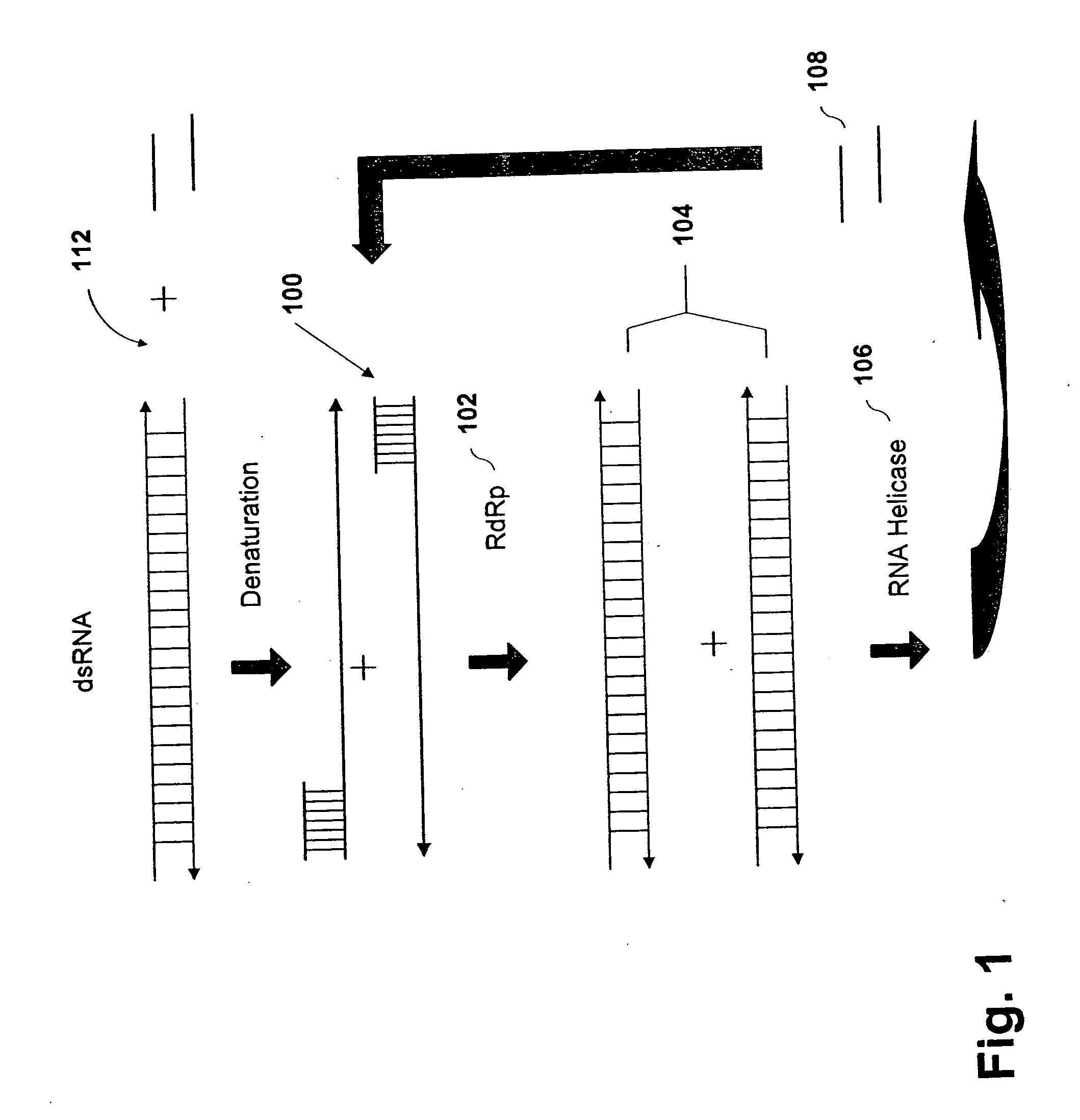
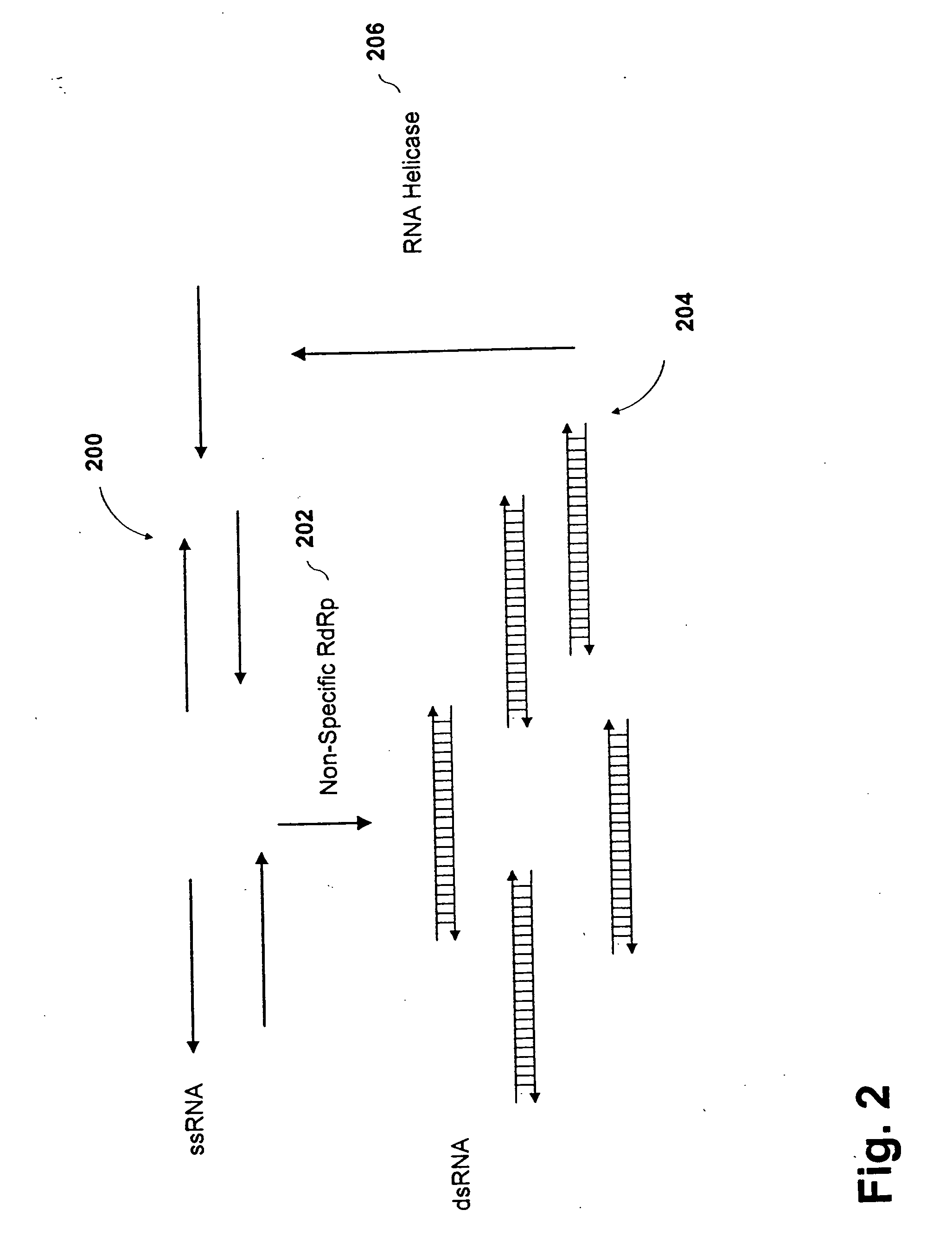
Methods and compositions for RNA amplification and detection using an RNA-dependent RNA-polymerase
Methods and compositions are provided for amplifying RNA from an RNA template. Methods and compositions are further provided for detecting an RNA in an RNA containing sample. Compositions used for the amplification of RNA from RNA include a RNA-dependent RNA-polymerase, a RNA helicase, and an energy source. Illustrative RNA-dependent RNA-polymerase enzymes are derived from the tomato or tobacco plant, while illustrative RNA helicase enzymes include the eIF4A and eIF4B proteins.
Owner:QIAGEN NORTH AMERICAN HLDG INC
Isothermal exponential RNA amplification in complex mixtures
Methods and compositions are provided for performing isothermal amplification of a nucleic acid target employing probes characterized by having a masked RNA polymer promoter unable to bind to a complementary initiator oligonucleotide and RNA polymerase and initiate transcription, a dsDNA sequence which when invaded by the target nucleic acid exposes the masked promoter to initiate transcription, and a template sequence, a portion of which is normally included in the dsDNA region, which when copied produces a product that can reinitiate the process of invading the dsDNA region and initiating transcription of another copy.
Owner:DISCOVERX INC
Anti-tumor T cell as well as preparation method and anti-tumor drug thereof
InactiveCN104630145AMammal material medical ingredientsBlood/immune system cellsAbnormal tissue growthDendritic cell
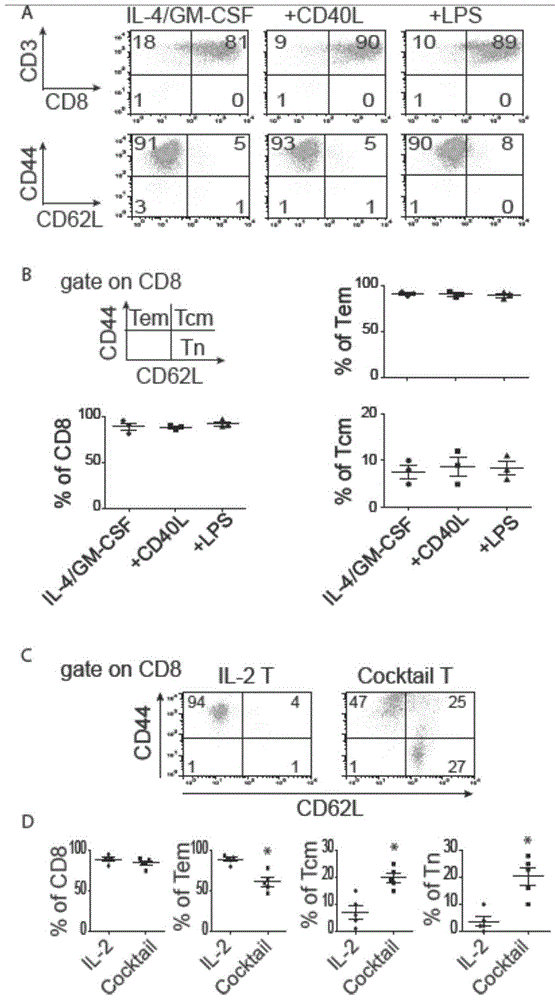
The invention provides a method for preparing an anti-tumor T cell in vitro through DC (dendritic cell, namely antigen presenting cell), wherein the anti-tumor T cell is a tumor total RNA-loaded DC amplification and enrichment anti-tumor T cell. The invention also provides a preparation method of the anti-tumor T cell and an anti-tumor drug or inoculation vaccine taking the anti-tumor T cell as an active ingredient. According to the invention, a specific anti-tumor T cell is amplified in vitro by taking the tumor total RNA-loaded DC as a platform, a small amount of tumor total RNA is amplified to an enough amount in vitro so as to realize clinical application, and the in-vivo anti-tumor effect in an IC (intracranial) tumor-bearing model is evaluated, including how the mature state of DC and the cell factor combination adjust the differentiation of the anti-tumor T cell as well as the synergistic anti-tumor effect thereof in a preclinical experiment model.
Owner:深圳市中美康士生物科技有限公司
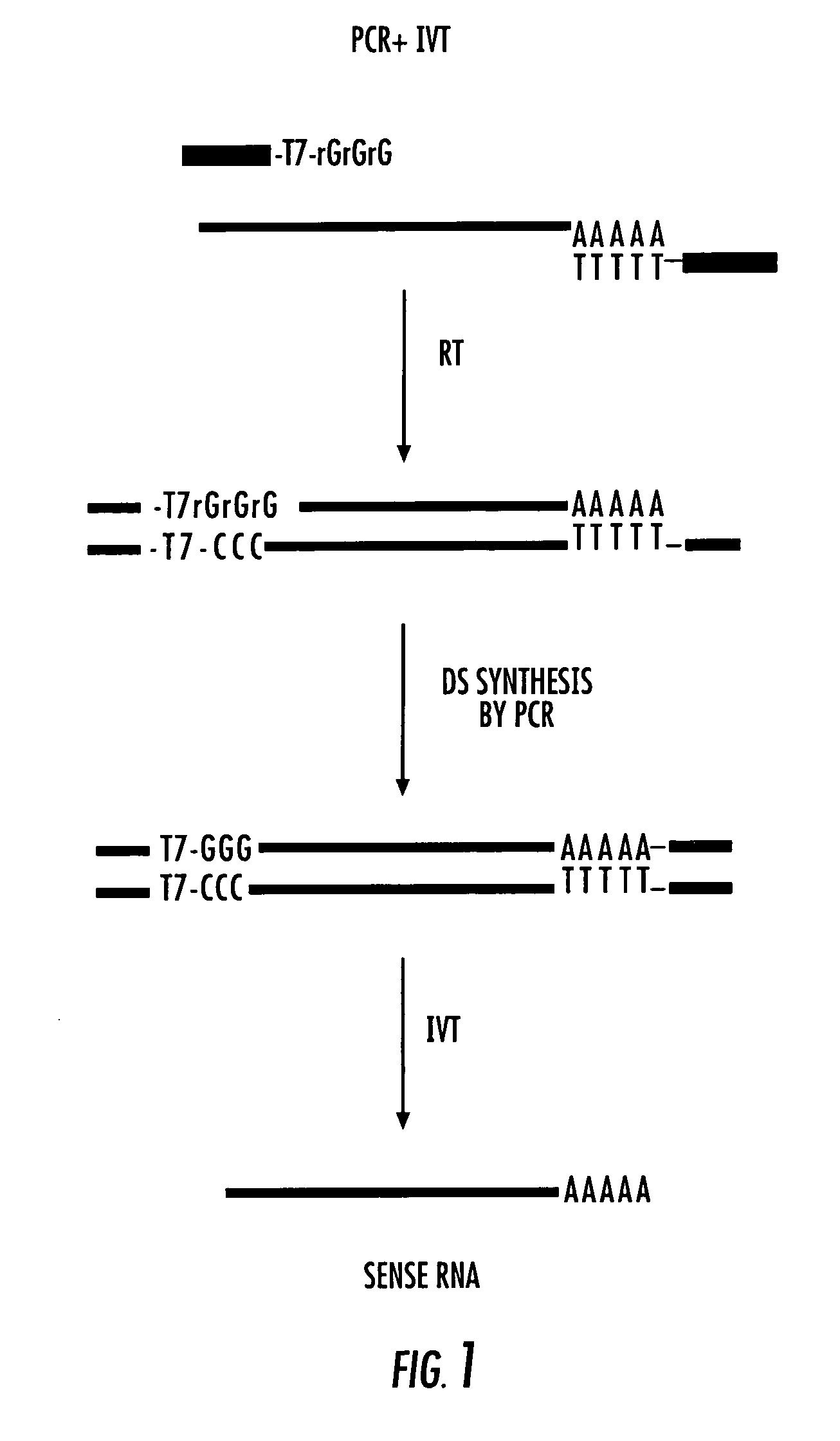
Random-primed reverse transcriptase-in vitro transcription method for rna amplification
InactiveUS20040081978A1Improve representationImprove abilitiesMicrobiological testing/measurementFermentationReverse transcriptaseSingle strand
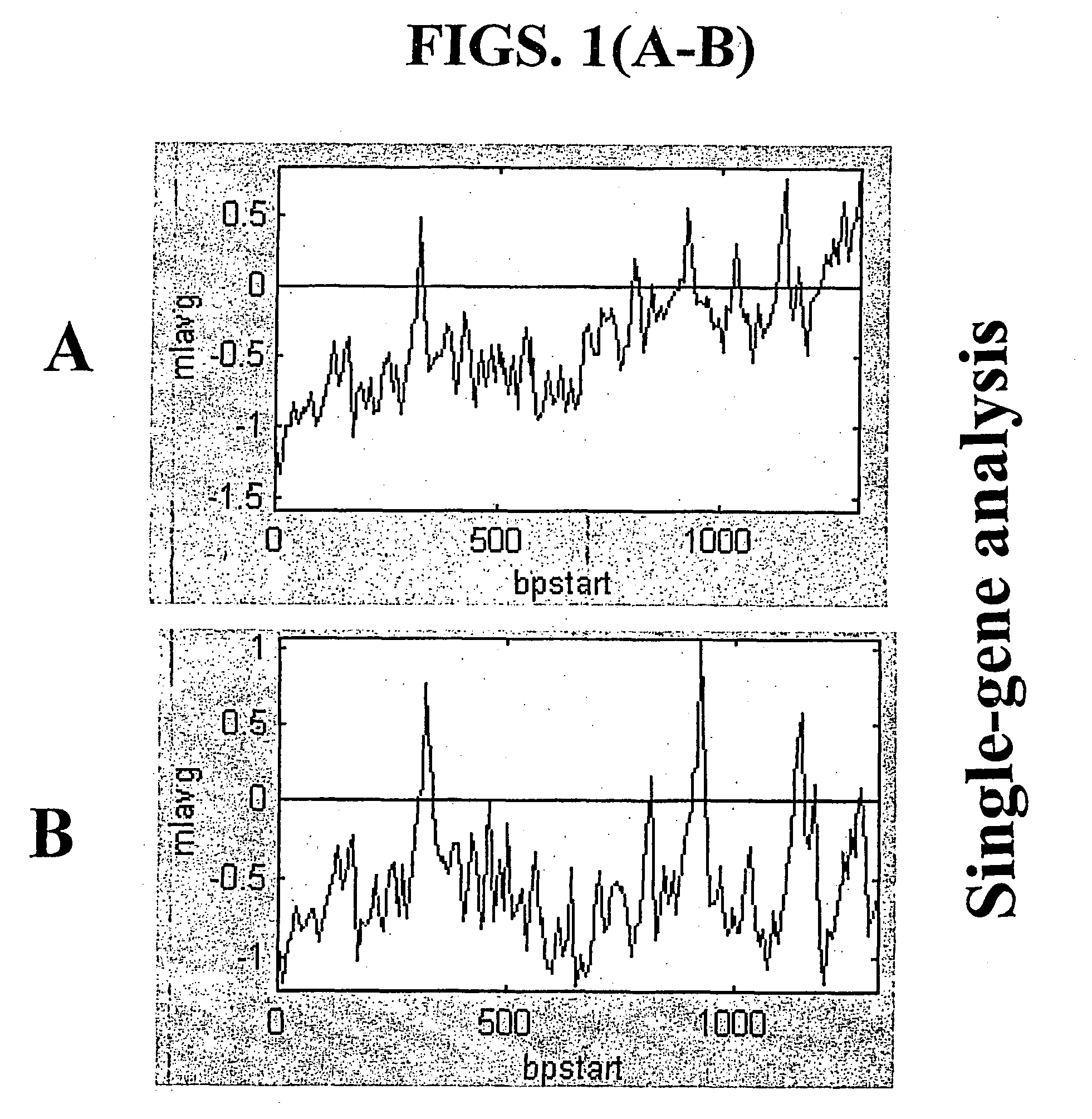
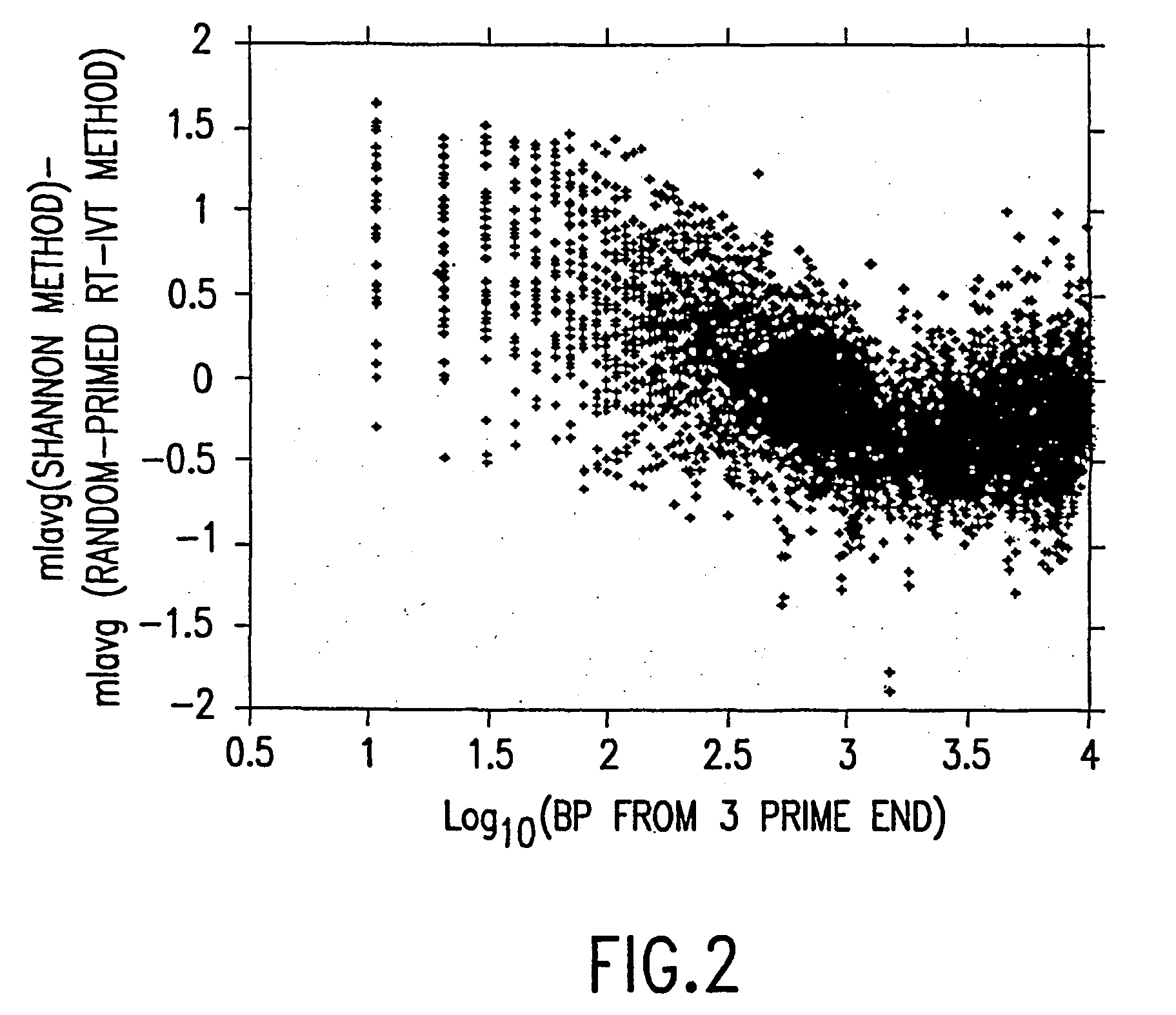
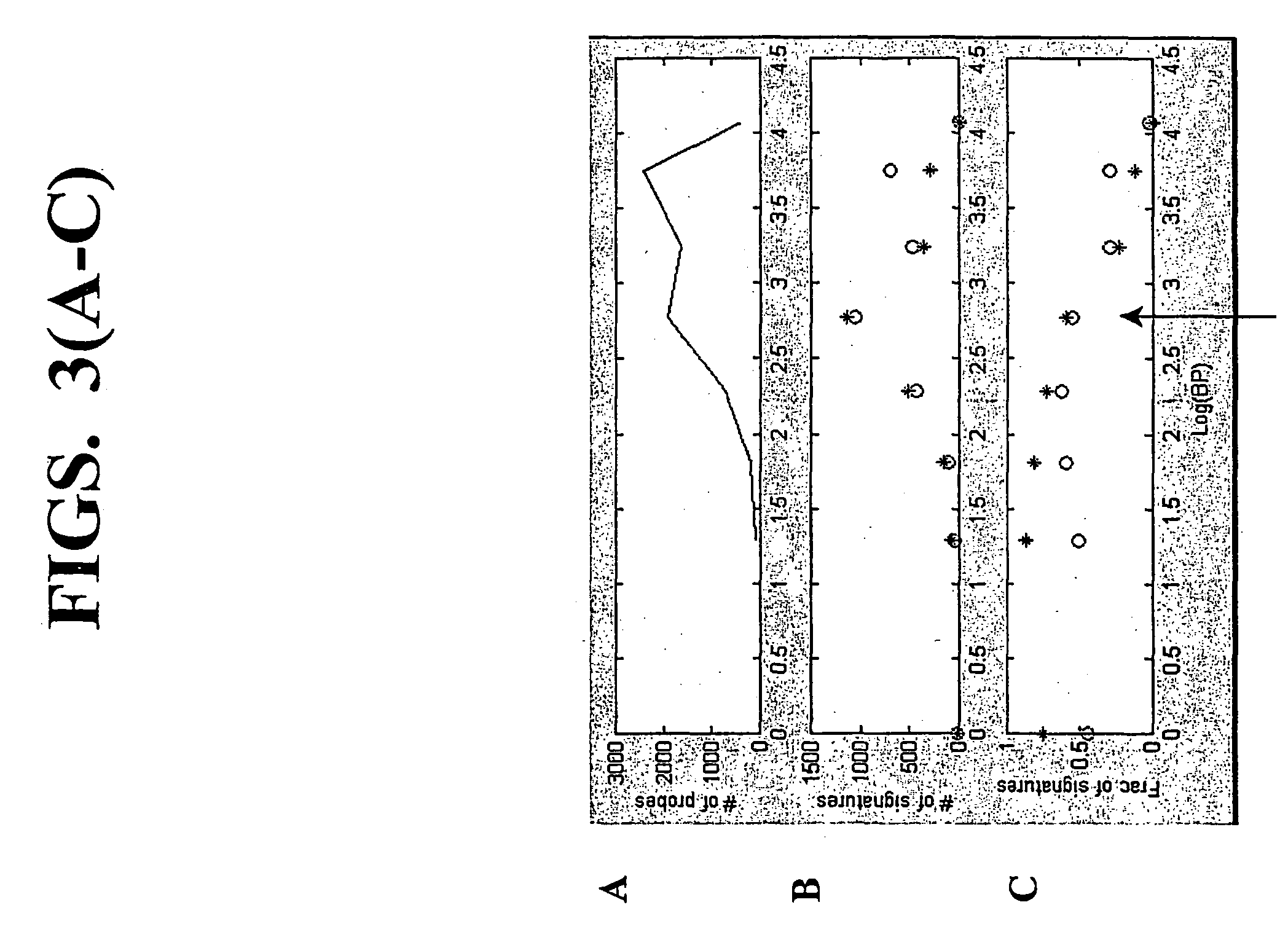
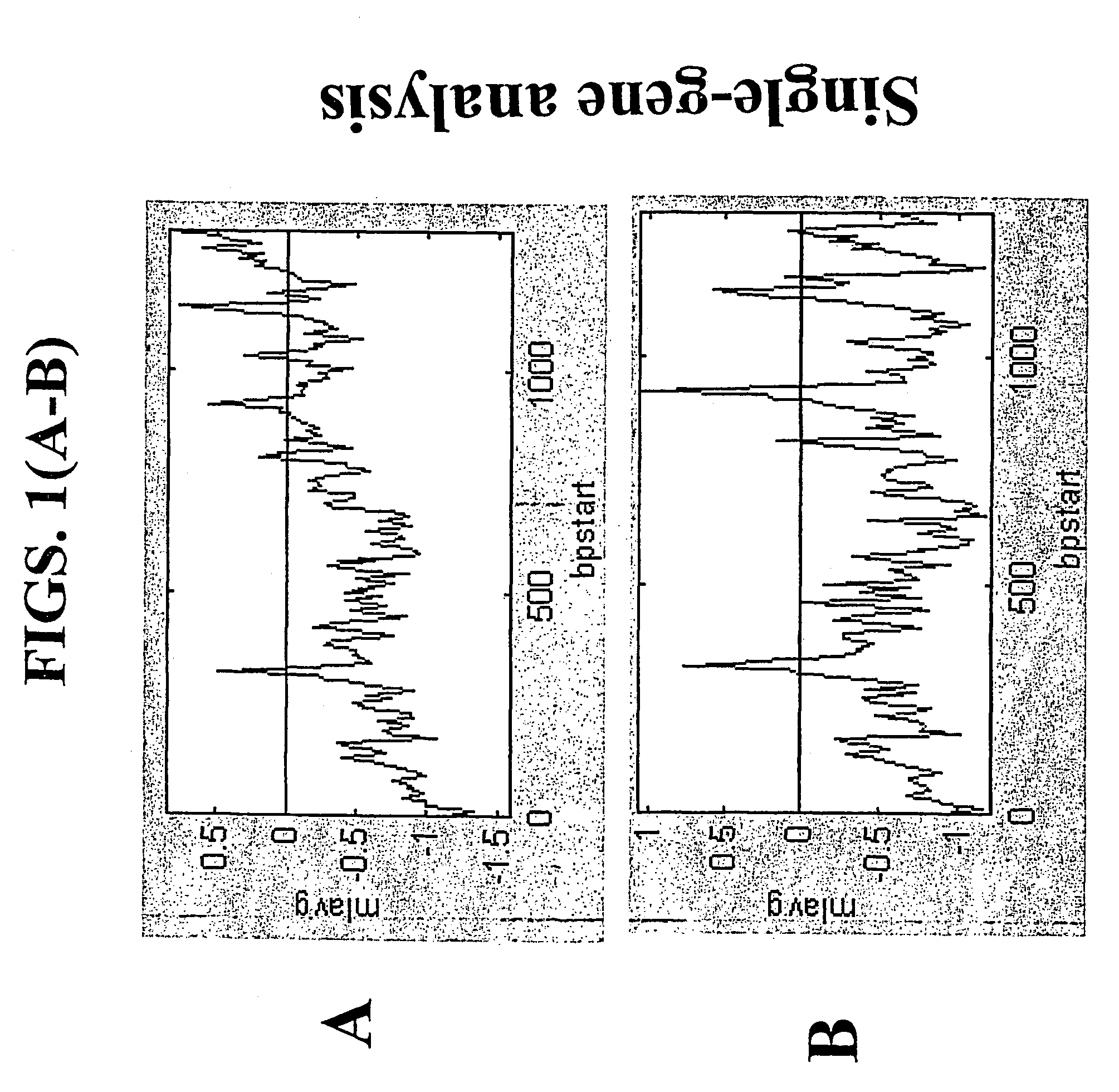
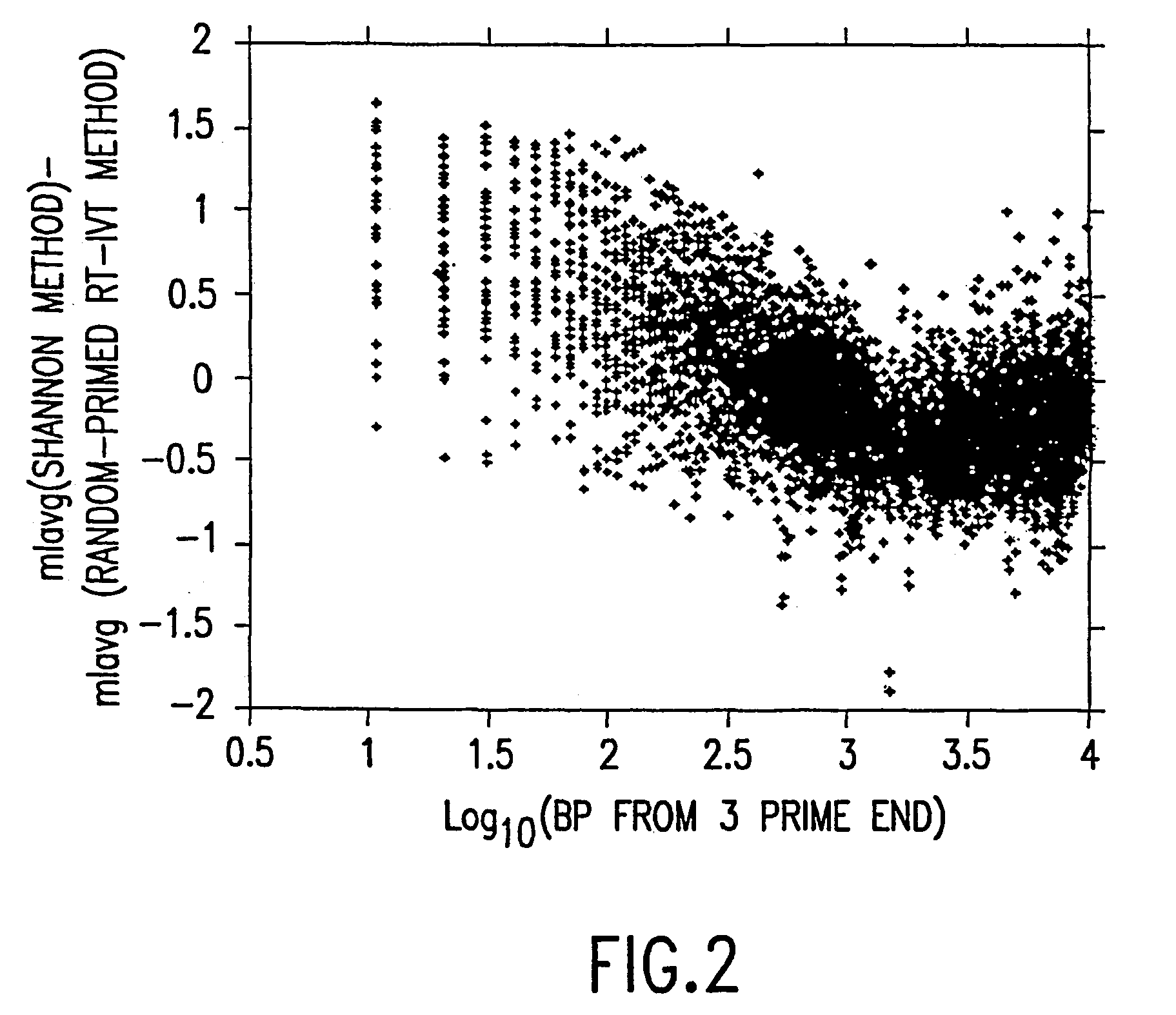
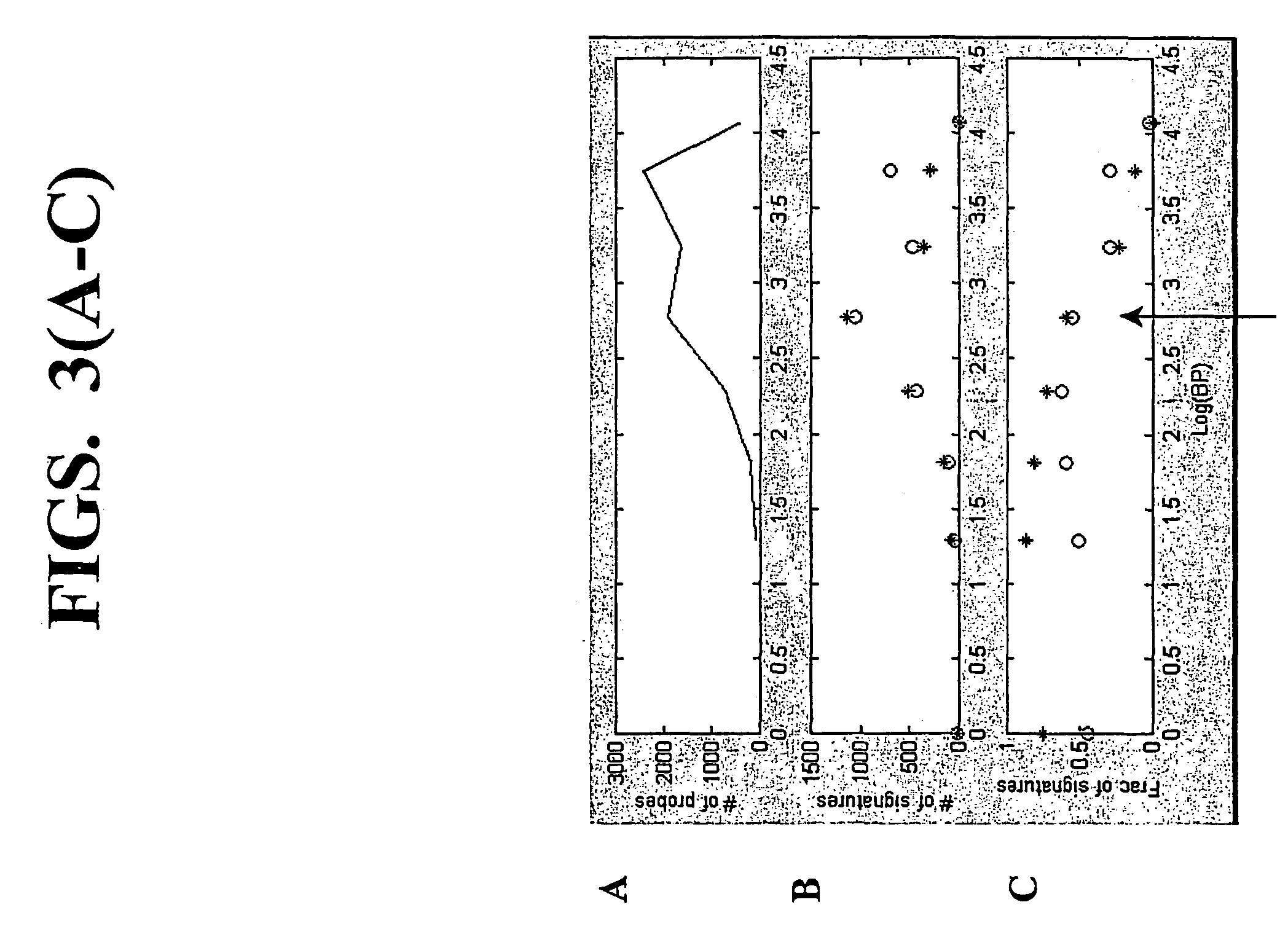
A random-primed reverse transcriptase-in vitro transcription method of linearly amplifying RNA is provided. According to the methods of the invention, source RNA (or other single-stranded nucleic acid), preferably, mRNA, is converted to double-stranded cDNA using two random primers, one of which comprises a RNA polymerase promoter sequence ("promoter-primer"), to yield a double-stranded cDNA that comprises a RNA polymerase promoter that is recognized by a RNA polymerase. Preferably, the primer for first-strand cDNA synthesis is a promoter-primer and the primer for second-strand cDNA synthesis is not a promoter-primer. The double-stranded cDNA is then transcribed into RNA by the RNA polymerase, optimally in the presence of a reverse transcriptase that is rendered incapable of RNA-dependent DNA polymerase activity during this transcription step. The subject methods produce linearly amplified RNA with little or no 3' bias in the sequences of the nucleic acid population amplified.
Owner:LIFE TECH CORP
Random-primed reverse transcriptase-in vitro transcription method for RNA amplification
InactiveUS7229765B2Improve abilitiesImprove representationMicrobiological testing/measurementFermentationReverse transcriptaseSingle strand
A random-primed reverse transcriptase-in vitro transcription method of linearly amplifying RNA is provided. According to the methods of the invention, source RNA (or other single-stranded nucleic acid), preferably, mRNA, is converted to double-stranded cDNA using two random primers, one of which comprises a RNA polymerase promoter sequence (“promoter-primer”), to yield a double-stranded cDNA that comprises a RNA polymerase promoter that is recognized by a RNA polymerase. Preferably, the primer for first-strand cDNA synthesis is a promoter-primer and the primer for second-strand cDNA synthesis is not a promoter-primer. The double-stranded cDNA is then transcribed into RNA by the RNA polymerase, optimally in the presence of a reverse transcriptase that is rendered incapable of RNA-dependent DNA polymerase activity during this transcription step. The subject methods produce linearly amplified RNA with little or no 3′ bias in the sequences of the nucleic acid population amplified.
Owner:LIFE TECH CORP
Novel microarray techniques for nucleic acid expression analyses
ActiveUS20060078925A1Eliminate biasConvenient experimentMicrobiological testing/measurementFermentationMiRNA GeneExonuclease I
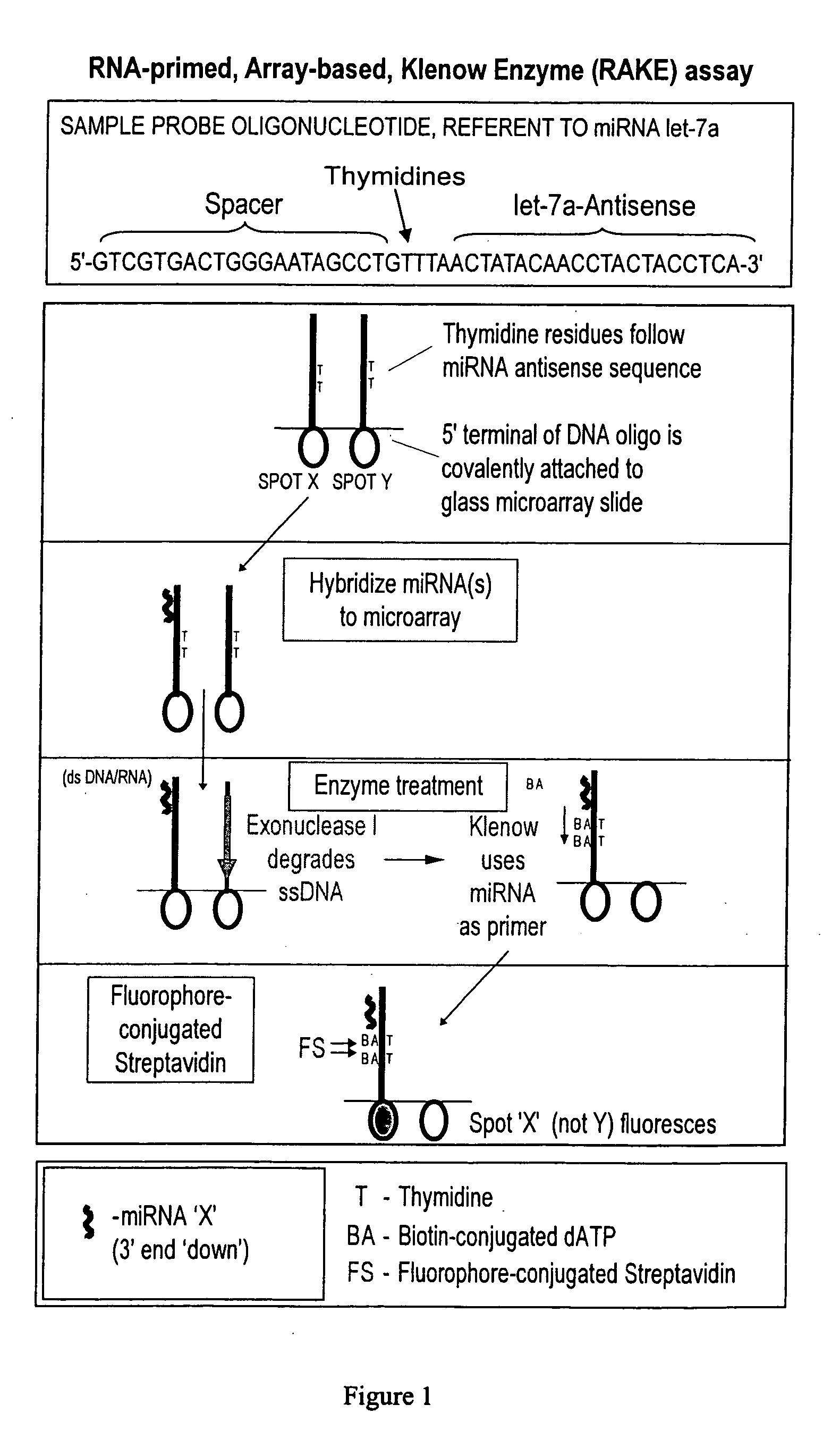
Provided are DNA microarray techniques that allow hybridization without RNA amplification, without using cDNA, and without labeling the nucleic acid prior to hybridization. Referred to as the Double-stranded Exonuclease Protection (DEP) assay, the technique permits the sample RNA to be used directly for hybridization, without manipulation in any way. Further provided is a microarray technique for high-throughput miRNA gene expression analyses, termed the RNA-primed, Array-based, Klenow Enzyme (RAKE) assay. The RAKE assay is a sensitive and specific technique for assessing single-stranded DNA and RNA targets, and offers specific advantages over Northern blots.
Owner:THE TRUSTEES OF THE UNIV OF PENNSYLVANIA
Methods of RNA amplification in the presence of DNA
InactiveUS7846666B2Microbiological testing/measurementFermentationReverse transcriptaseDNA Polymerase Inhibitor
The invention provides methods for amplification of RNA. The methods are particularly suitable for specifically amplifying RNA in the presence of DNA. The methods involve producing a marked first primer extension product from a target RNA in the presence of a DNA-dependent DNA polymerase inhibitor, which prevents replication of DNA by the reverse transcriptase enzyme. The marked nucleic acid products are subsequently selectively amplified in the presence on non-marked nucleic acids. The methods are useful for production and analysis of polynucleotide sequences complementary to an RNA sequence. The methods are useful for preparation of nucleic acid libraries and substrates for analysis of gene expression of cells in biological samples. The invention also provides compositions and kits for practicing the amplification methods, as well as methods which use the amplification products.
Owner:NUGEN TECH
Nucleic acid detection method combining RNA amplification with hybrid capture method
InactiveCN103525942AEnable high-throughput detectionNot easy to inactivateMicrobiological testing/measurementBiotin-streptavidin complexStreptolydigin
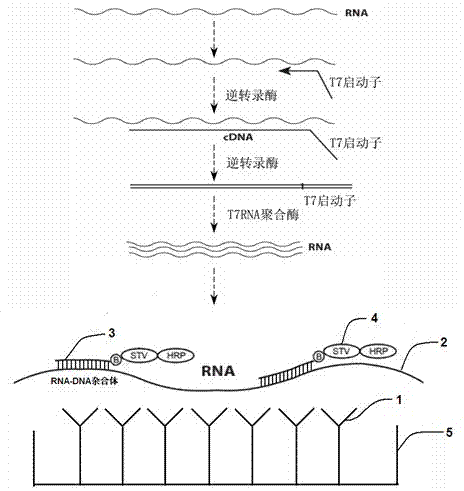
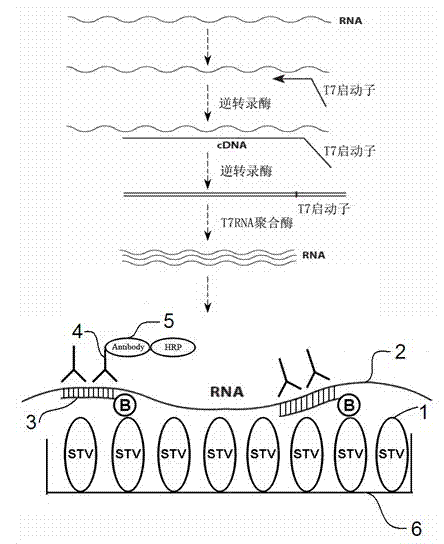
The invention relates to a nucleic acid detection method combining RNA amplification with a hybrid capture method. The nucleic acid detection method includes the following steps of extracting nucleic acid from a sample or splitting the sample to release the nucleic acid to be detected, conducting transcription and amplification on the nucleic acid to be detected to obtain an RNA product, adding the obtained RNA product together with a DNA probe marked with biotins to a solid phase so that hybridization can be conducted, forming a DNA / RNA heterozygote through hybridization, coating the solid phase with specific antibodies for recognizing the heterozygote or avidin or streptavidin, capturing the heterozygote through the solid phase, and adding avidin or streptavidin or antibodies for recognizing the DNA / RNA heterozygote to the solid phase to achieve obtaining of a nucleic acid signal, wherein the avidin or the streptavidin or the antibodies for recognizing the DNA / RNA heterozygote are marked with enzymes with the secondary amplification capacity. According to the nucleic acid detection method, the inherent pollution problem of the PCR is avoided, RNA molecules can be directly detected, inverse transcription and pre-dissociation of RNA do not need to be conducted, and the operation steps are greatly simplified.
Owner:武汉中帜生物科技股份有限公司
Methods for PCR and HLA typing using raw blood
Owner:GENOMICS USA
Fast detection method of nucleic acid of A H1N1 influenza virus and kit thereof
InactiveCN101845517AIncreased sensitivitySolve pollutionMicrobiological testing/measurementFluorescence/phosphorescenceSingle-Stranded RNAReverse transcriptase
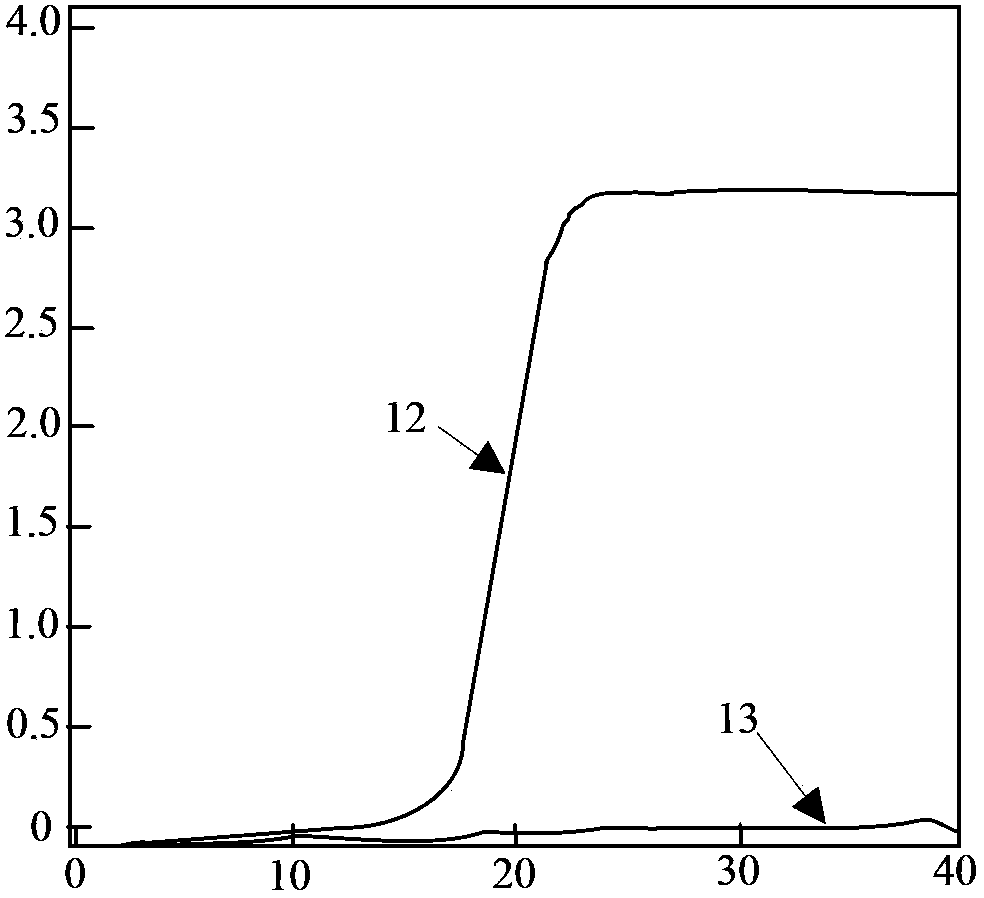
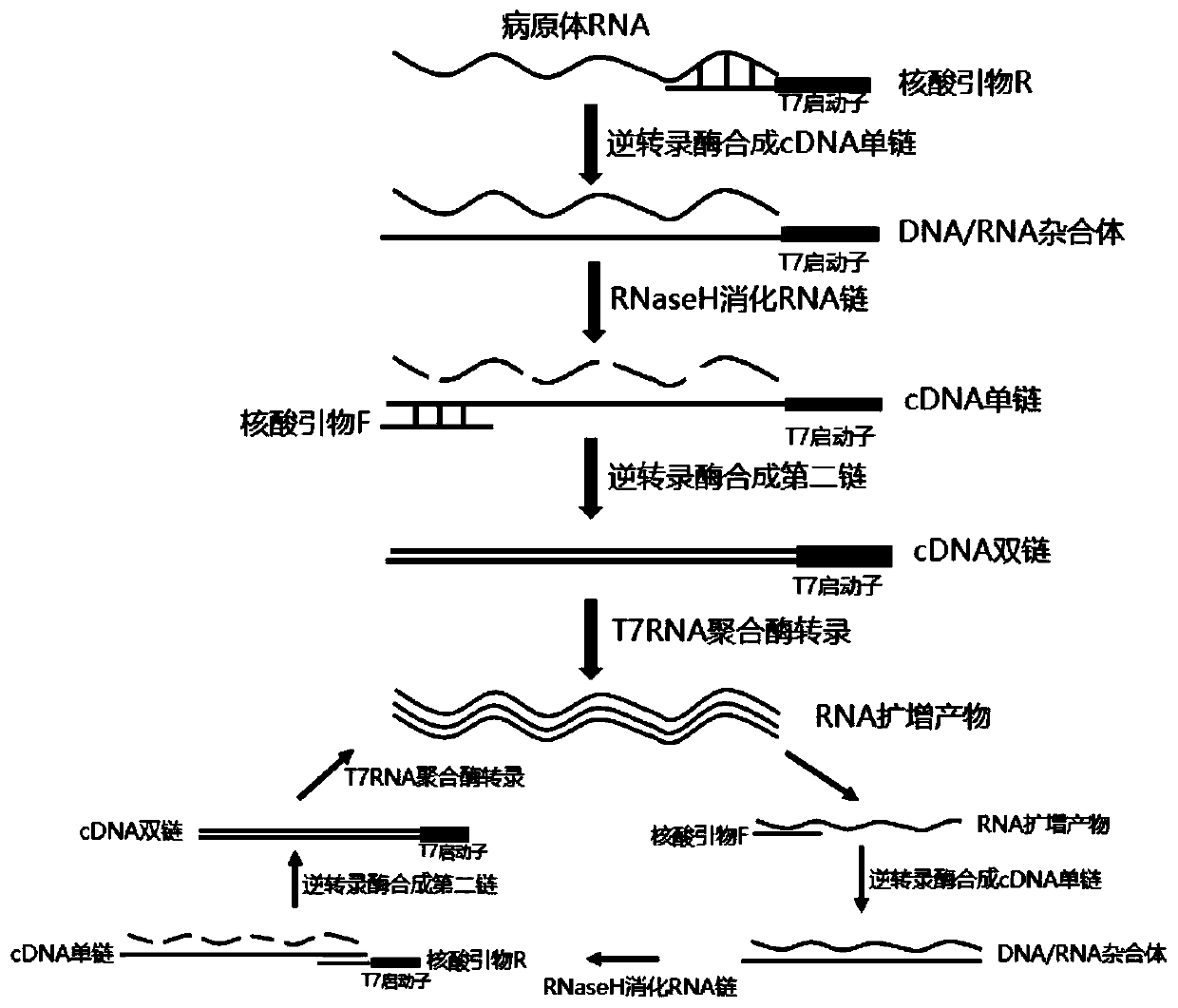
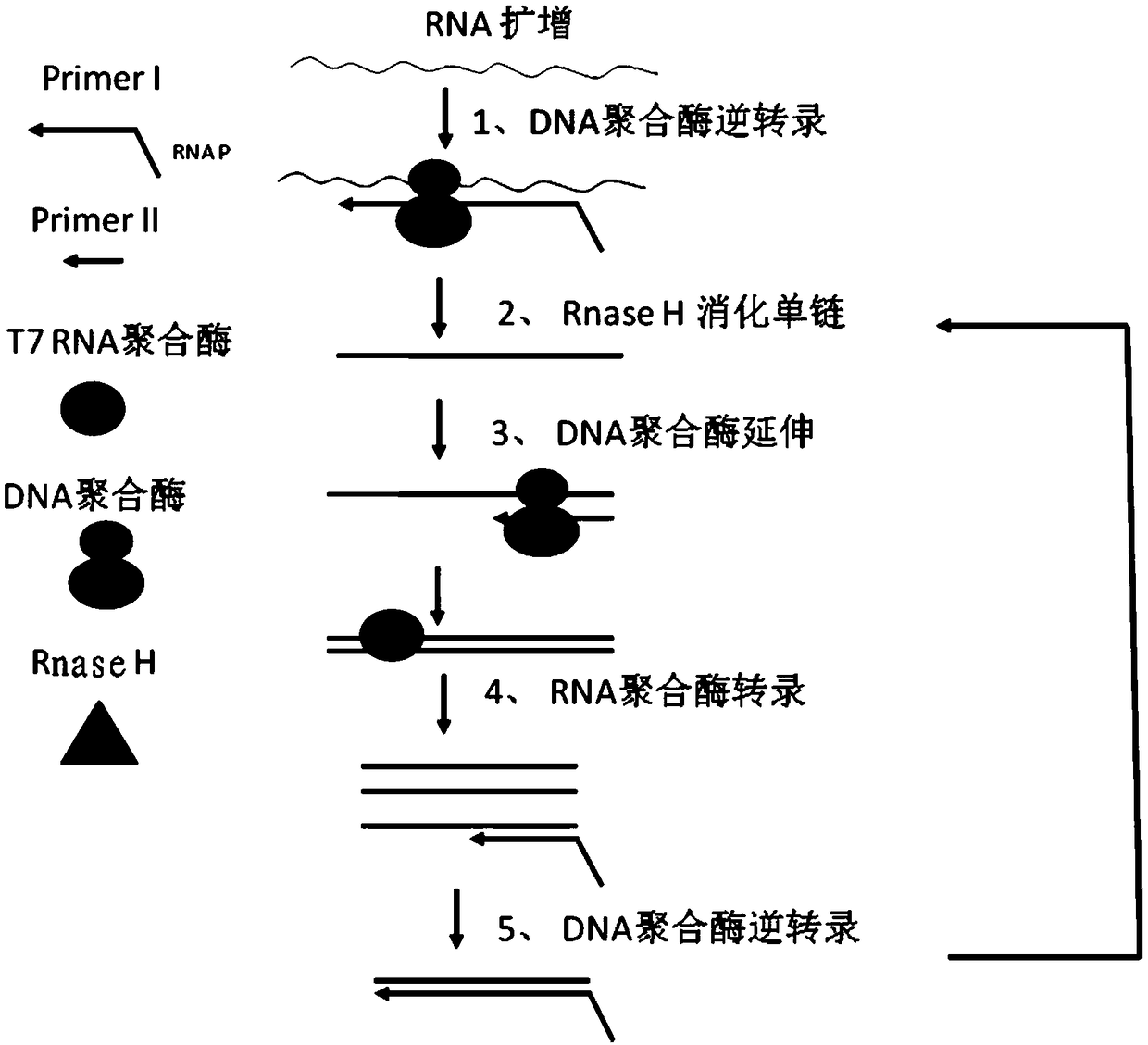
The invention relates to a fast detection method of nucleic acid of A H1N1 influenza virus, and a kit thereof, belonging to the technical field biotechnology. The invention provides a fast and synthermal RNA amplification method of A H1N1 influenza virus, and the accomplishment of the entire reaction depends on the cooperation of the AMV reverse transcriptase, the T7RNA polymerase and nuclease H (RNaseH), without the needs of the special instruments and the temperature cycling, the amplification efficiency is larger than or equal to that of the PCR, and the reaction product is single stranded RNA. The invention has the advantages of having equal or higher sensibility than that of the RT-PCR detection method, avoiding the pollution problem of the RT-PCR amplification product due to that the amplification product is RNA, finishing the reaction in one hour, and simple and convenient operation. The reaction can be monitored in real time and detected by the end-point method, can be used as the important tool in the detection of A H1N1 influenza virus.
Owner:JIANGSU PROVINCIAL CENT FOR DISEASE PREVENTION & CONTROL
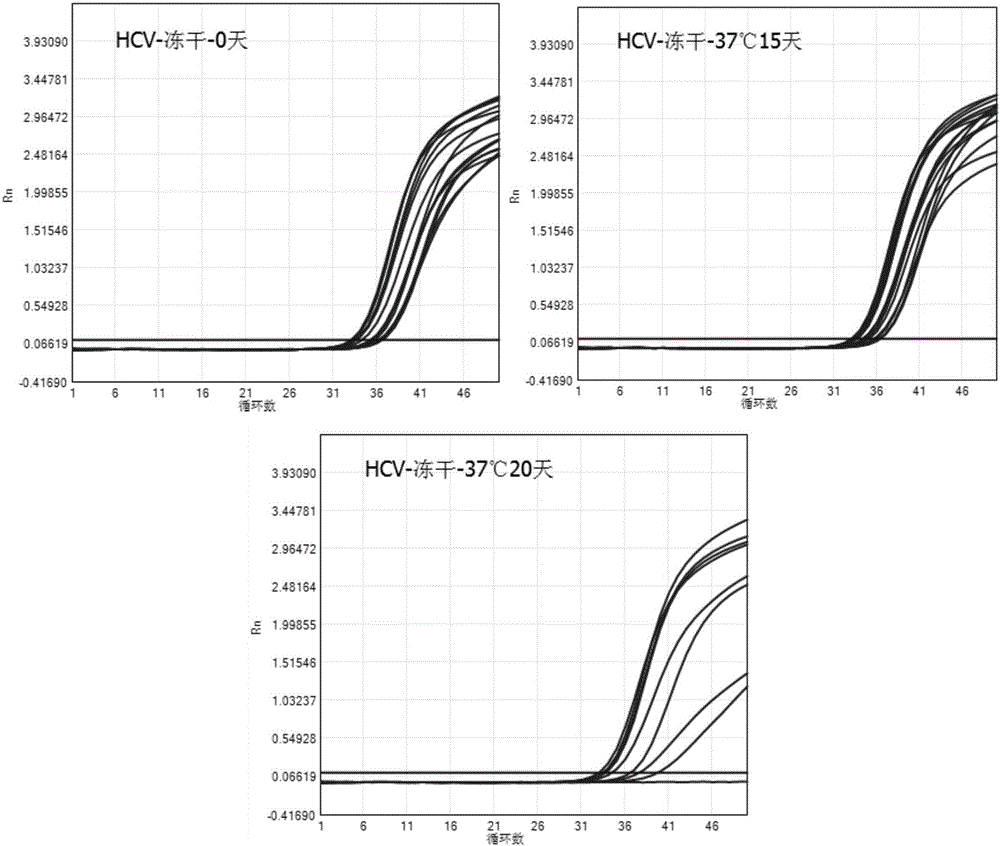
Freeze-drying protective agent and freeze-drying method for RNA amplification reaction agent
InactiveCN106591432ALow costImprove convenienceMicrobiological testing/measurementSucroseFreeze-drying
The invention discloses a freeze-drying protective agent and a freeze-drying method for an RNA amplification reaction agent, belonging to the field of biotechnologies. The freeze-drying protective agent contains mycose, polysucrose and water, and is prepared by mixing 6.4-16.0 g of mycose and 0.4-1.0 g of polysucrose and adding with water till the volume is 40ml. The invention further provides the freeze-drying method for the RNA amplification reaction agent. The freeze-drying method comprises the following steps: a, uniformly mixing the freeze-drying protective agent and a real-time fluorescent RT-PCR reaction agent, thus obtaining real-time fluorescent RT-PCR reaction liquid; and b, carrying out freeze drying on the RT-PCR reaction liquid, thus obtaining the RT-PCR freeze-dried reagent. With the adoption of the freeze-drying protective agent and the freeze-drying method provided by the invention, the RNA fluorescent quantitative PCR reaction agent can be stably stored for a long time at 2-8 DEG C or even at the room temperature.
Owner:ZHUHAI LIVZON DIAGNOSTICS
Freeze-drying protective agent and freeze-drying method of RNA amplification reaction reagent
ActiveCN111560417AGuaranteed stabilityGood freeze-drying effectMicrobiological testing/measurementBiotechnologySucrose
The invention discloses a freeze-drying protective agent and a freeze-drying method of an RNA amplification reaction reagent, and relates to the technical field of biology. The freeze-drying protective agent is prepared from the following components: 5 to 20 percent of trehalose, 5 to 15 percent of mannitol, 0.5 to 5mg / mL of bovine serum albumin, 0.05 to 0.5 percent of a surfactant and 0.01 to 0.05 percent of a defoaming agent. In the implementation process, a small amount of lentinan and cane sugar are added into a freeze-dried reagent; it is found that after addition, the freeze-drying effect of the freeze-dried reagent can be effectively improved, the freeze-dried reagent can be stored for a long time under the normal temperature condition by controlling the concentration ratio of trehalose to lentinan to sucrose, and the freeze-driedreagent still has high sensitivity after being stored for 6 months under the room temperature condition.
Owner:INTEGRATED BIOSYSTEMS CO LTD +1
Primer for amplifying short-chain RNA (ribonucleic acid) and related method thereof
InactiveCN103509789ASimple designEasy to synthesizeMicrobiological testing/measurementDNA preparationMicrobiologyNucleotide sequencing
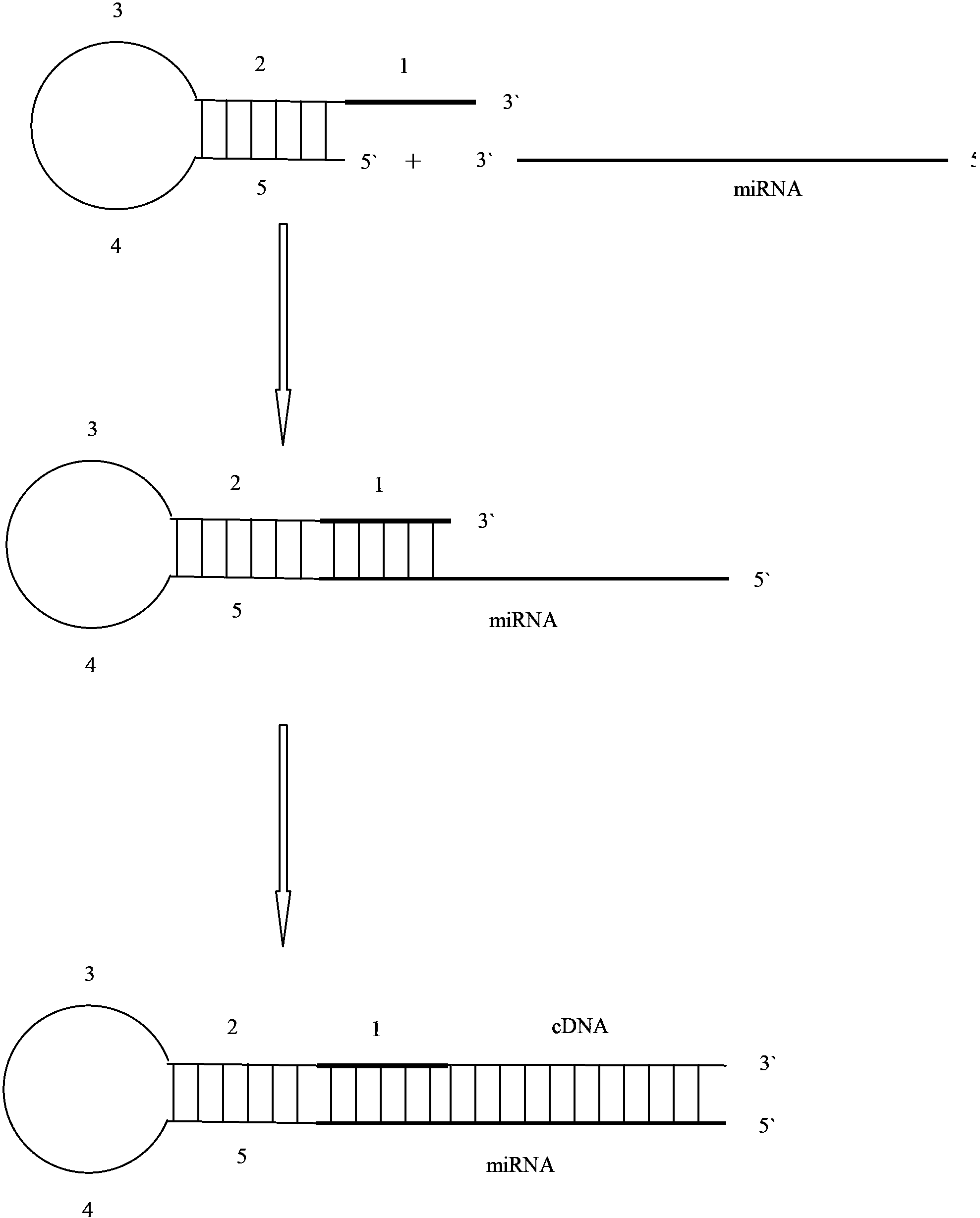
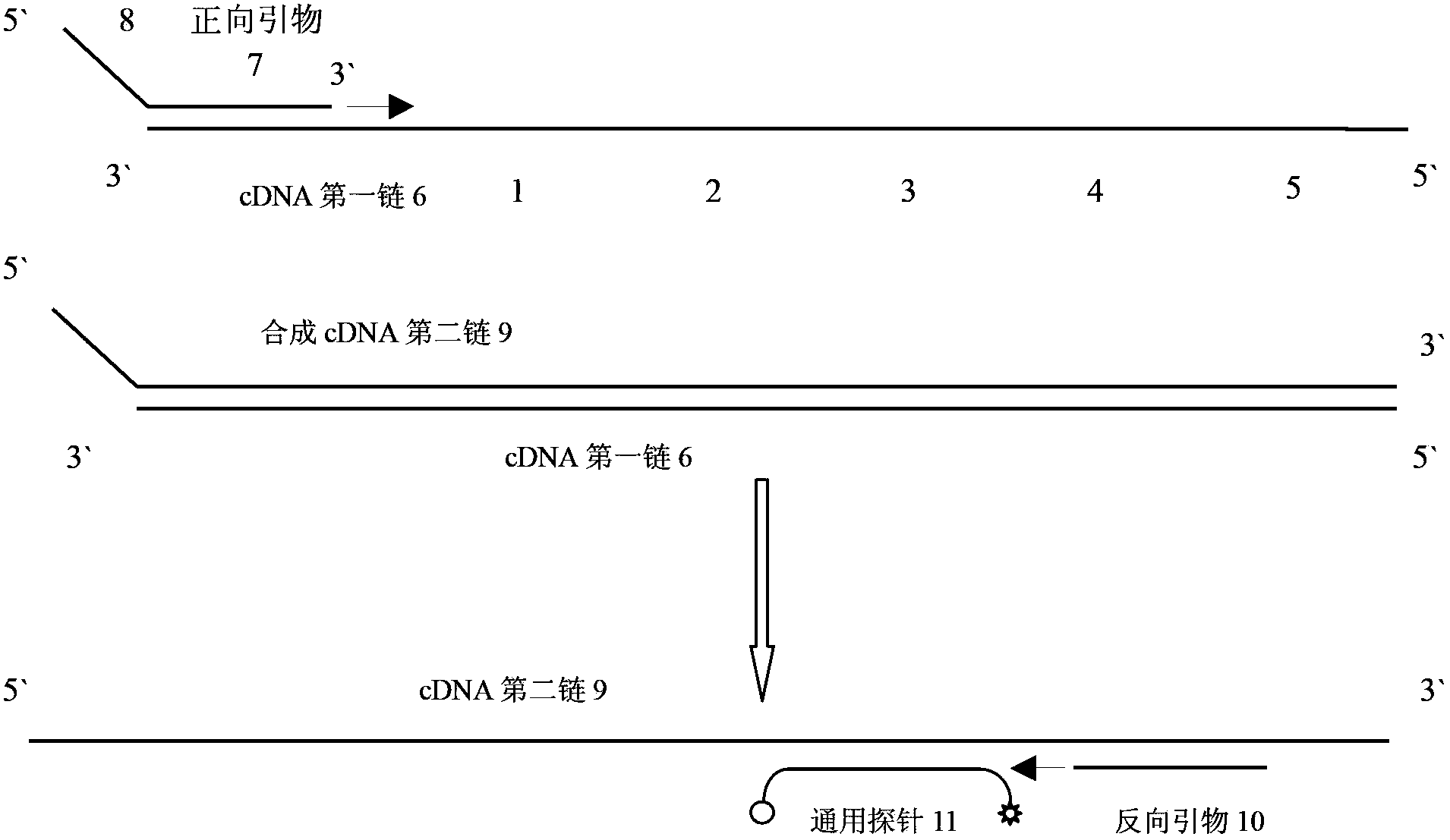
The invention discloses a primer for amplifying a short-chain RNA (ribonucleic acid) and a related method thereof. The primer is oligonucleotide; a fragment of nucleotide sequence at the 5' end of the primer is fixed, and forms a structure with a nucleotide loop and a nucleotide stem; the 3' end of the primer is connected with 6 to 8 nucleotides, and is paired with the 3' end of a mature miR to form specific complementary binding; the 3' end of the nucleotide loop contains a fragment of nucleotide sequence with GC content of over 70 percent, and the fragment of nucleotide sequence is called a universal probe region; nucleotides on the 8th to 30th sites at the 5' end of the primer form a universal reverse primer region. The primer has an internal double-chain structure, and cannot be bound to a specific sequence in a nucleotide chain under the action of steric hindrance, and the reverse transcription of the sequence is avoided; the primer is only specifically paired with and bound to the 3' end for specific reverse transcription. The primer is high in specificity, easy to design, convenient to synthesize and suitable for the reverse transcription of the short-chain RNA, especially the mature miR, and the formation of a primer dimer is avoided.
Owner:ZHOUSHAN HOSPITAL
Colloidal gold chromatography kit for nucleic acid detection of novel coronaviruses (2019-nCoV) and application thereof
ActiveCN111455099AEasy to degradeAvoid pollutionMicrobiological testing/measurementMicroorganism based processesRNA extractionNucleic acid detection
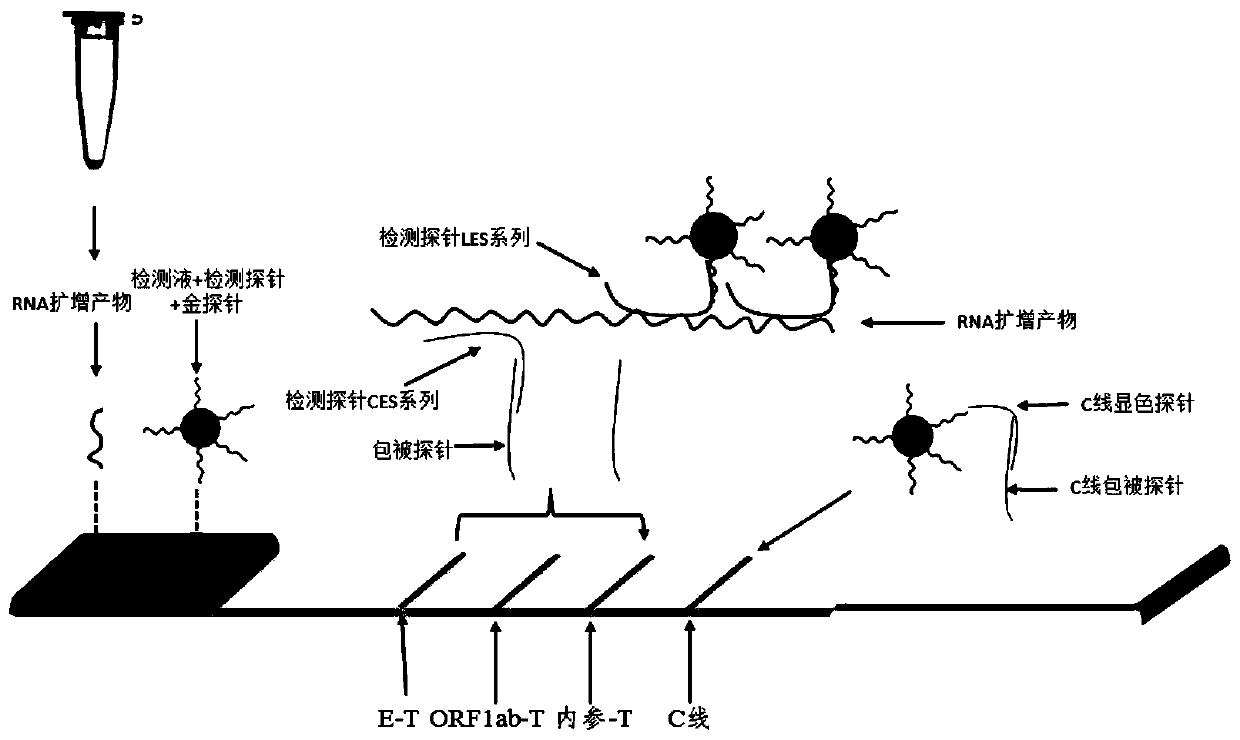
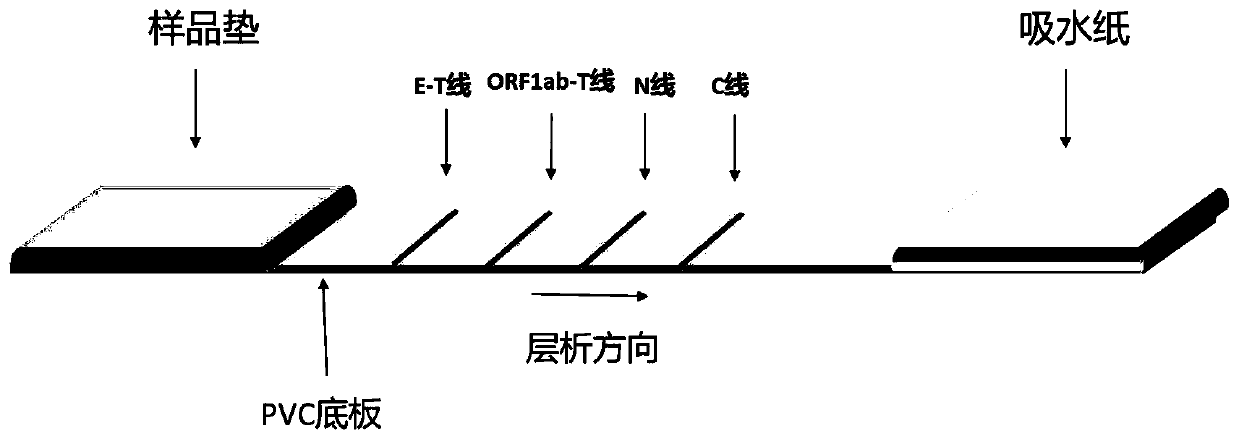
The invention discloses a colloidal gold chromatography kit for detecting novel coronaviruses (2019-nCoV) and an application thereof. According to the kit, a collected sample is cracked by a cell lysis solution to release pathogen nucleic acid, and then under the action of reverse transcriptase and T7RNA polymerase, amplification of pathogen nucleic acid fragments is realized through reverse transcription and transcription processes. An amplified RNA product is recognized and captured by a specific probe in a detection solution to form an RNA amplification product-specific probe-gold probe compound, and the compound is fixed on an NC membrane through lateral flow chromatography to form a visible strip, so that the detection of the pathogen nucleic acid is realized. The kit has no RNA extraction process, does not need a special instrument, has difficulty in pollution in actual detection based on RNA isothermal amplification, has the advantages of high sensitivity, high specificity and simplicity in operation, and makes wide application of novel coronavirus (2019-nCoV) nucleic acid detection possible.
Owner:武汉中帜生物科技股份有限公司
Method for detecting viable bacteria of Mycobacterium tuberculosis through isothermal amplification of nucleic acid and kit
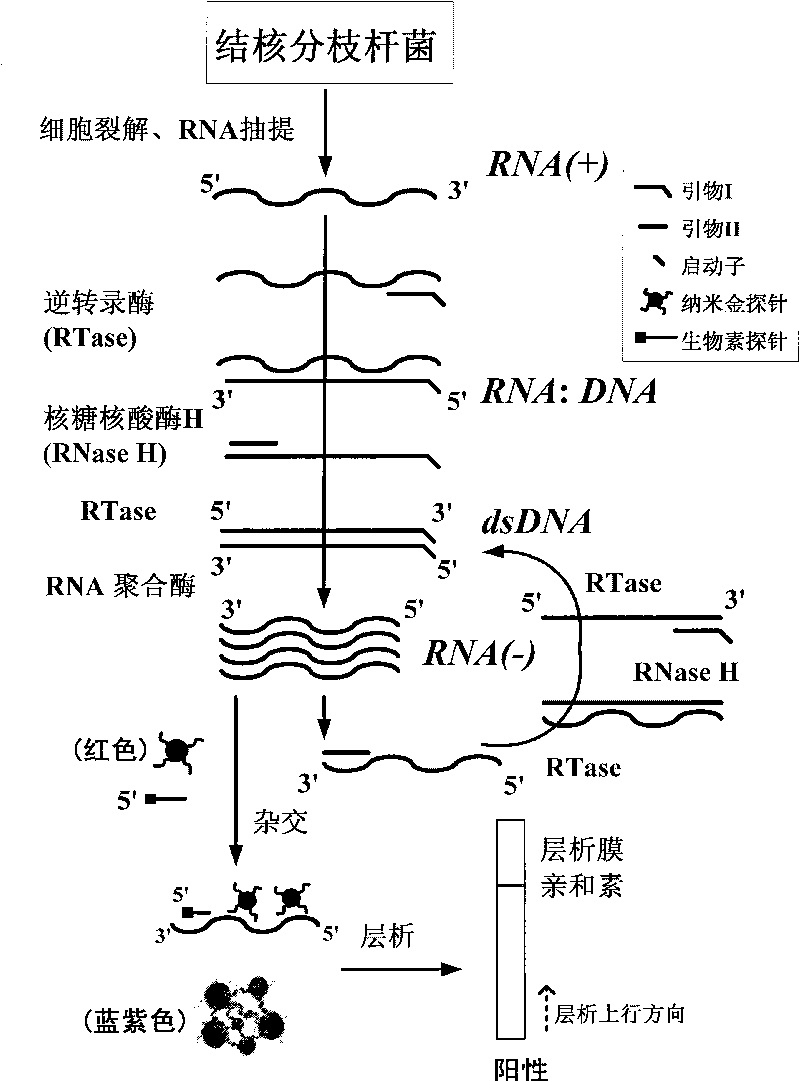
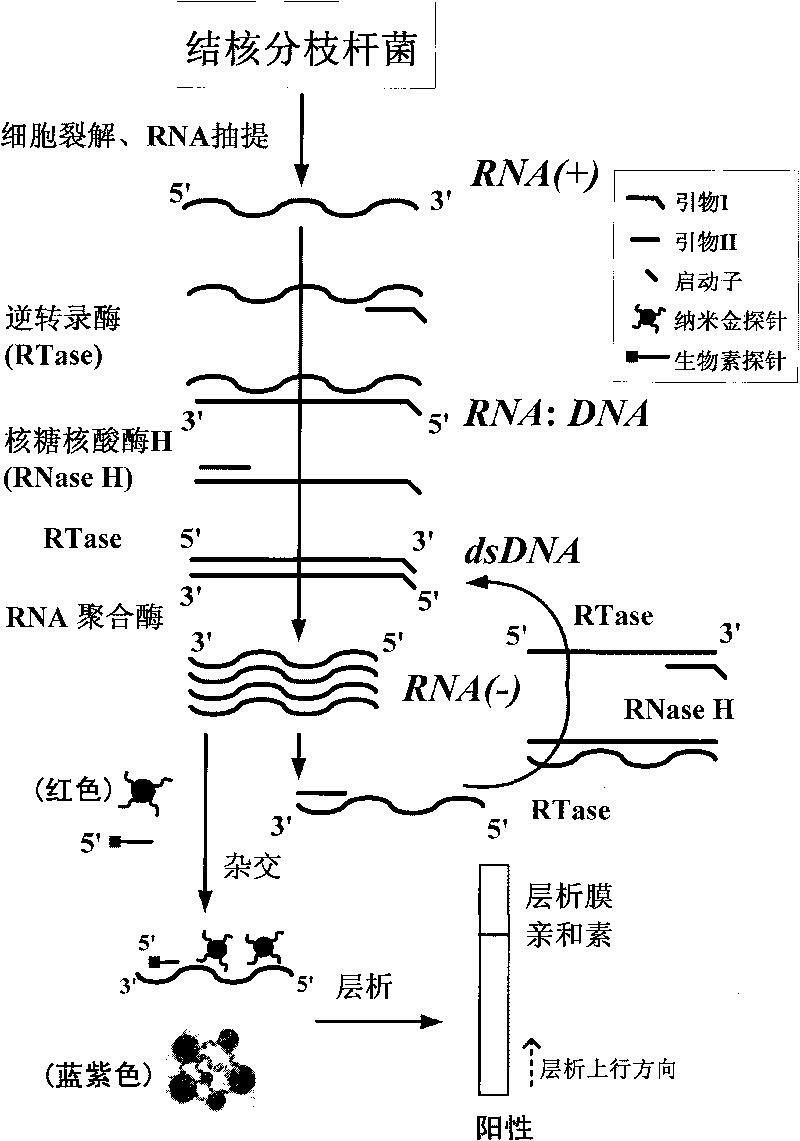
InactiveCN101736078AEasy and intuitive to observeEasy to excludeMaterial analysis by observing effect on chemical indicatorMicrobiological testing/measurementTreatment effectReverse transcriptase
The invention relates to a method for detecting viable bacteria of Mycobacterium tuberculosis through isothermal amplification of nucleic acid and a kit thereof. The method comprises the following detection steps: unlinking an mRNA template of the Mycobacterium tuberculosis at the temperature of between 60 and 70 DEG C and then reducing temperature; adding reverse transcriptase, RNaseH and RNA polymerase, and performing isothermal amplification at the temperature of between 37 and 42 DEG C under the guidance of a primer to obtain RNA amplicon; and then detecting the amplicon by utilizing a nanogold probe and a capture probe, and obtaining a detection result through chromatography hybridization color development reaction. That a color developing stripe appears on a hybrid membrane represents mRNA positive (the viable bacteria exist), no stripe represents negative. The kit adopting the method can detect the viable bacteria of the Mycobacterium tuberculosis, has the advantages of accuracy, sensitivity, simpleness, convenience, quickness and the like, can overcome the defects of long detection time, complex operation, low specificity and the like of the conventional methods, and can serve as an auxiliary experimental means for related researches such as diagnosis and prevention of tuberculosis, observation of treatment effect, screening of tuberculostatics, and sensitivity experiments.
Owner:SHANGHAI FOSUN PHARMA (GROUP) CO LTD +2
Identification of genes involved in fertility, ovarian function and/or fetal/newborn viability
InactiveUS20070054289A1Enhance their pregnancy competencyGood curative effectMicrobiological testing/measurementFermentationDiseaseHormone function
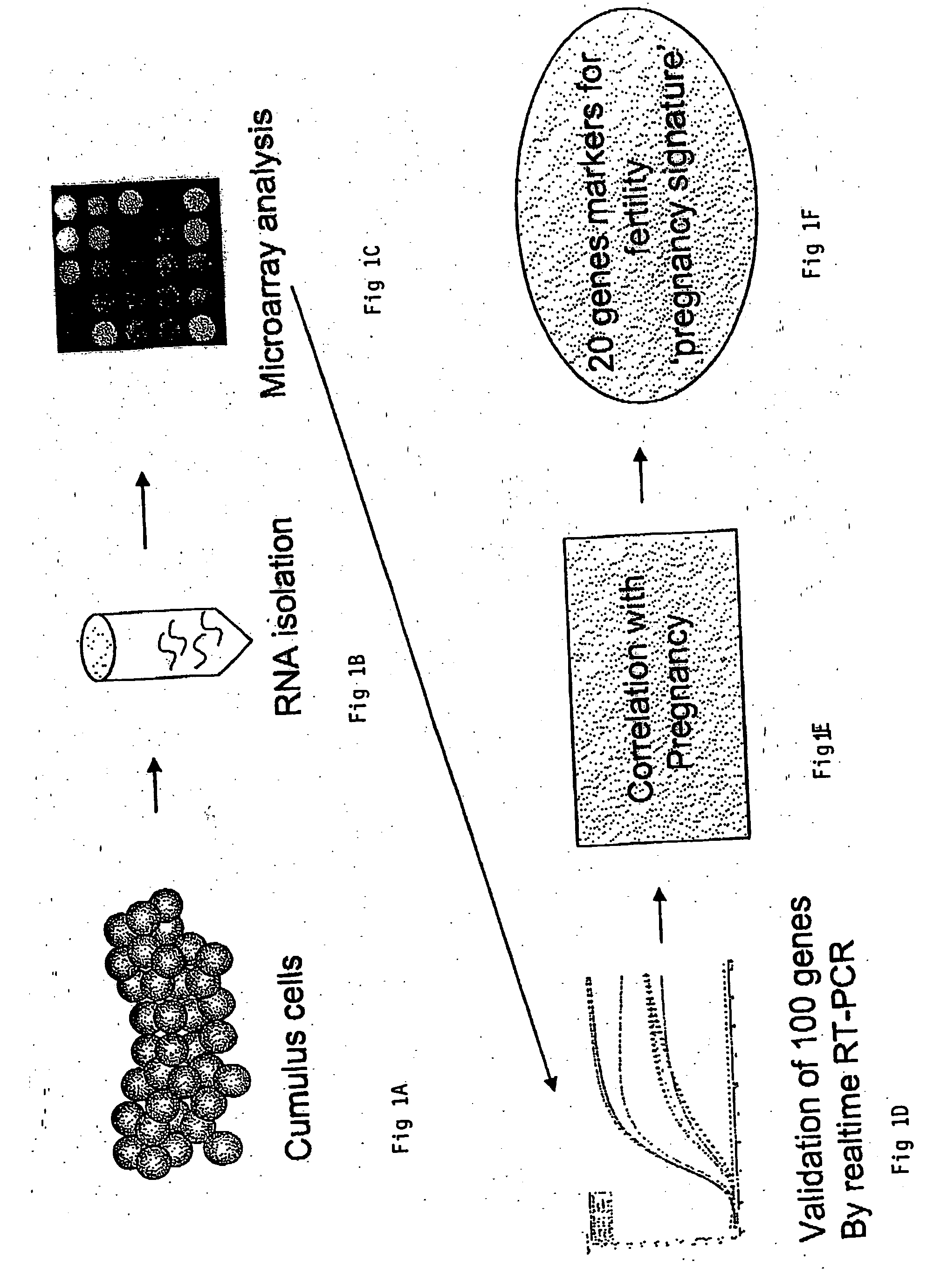
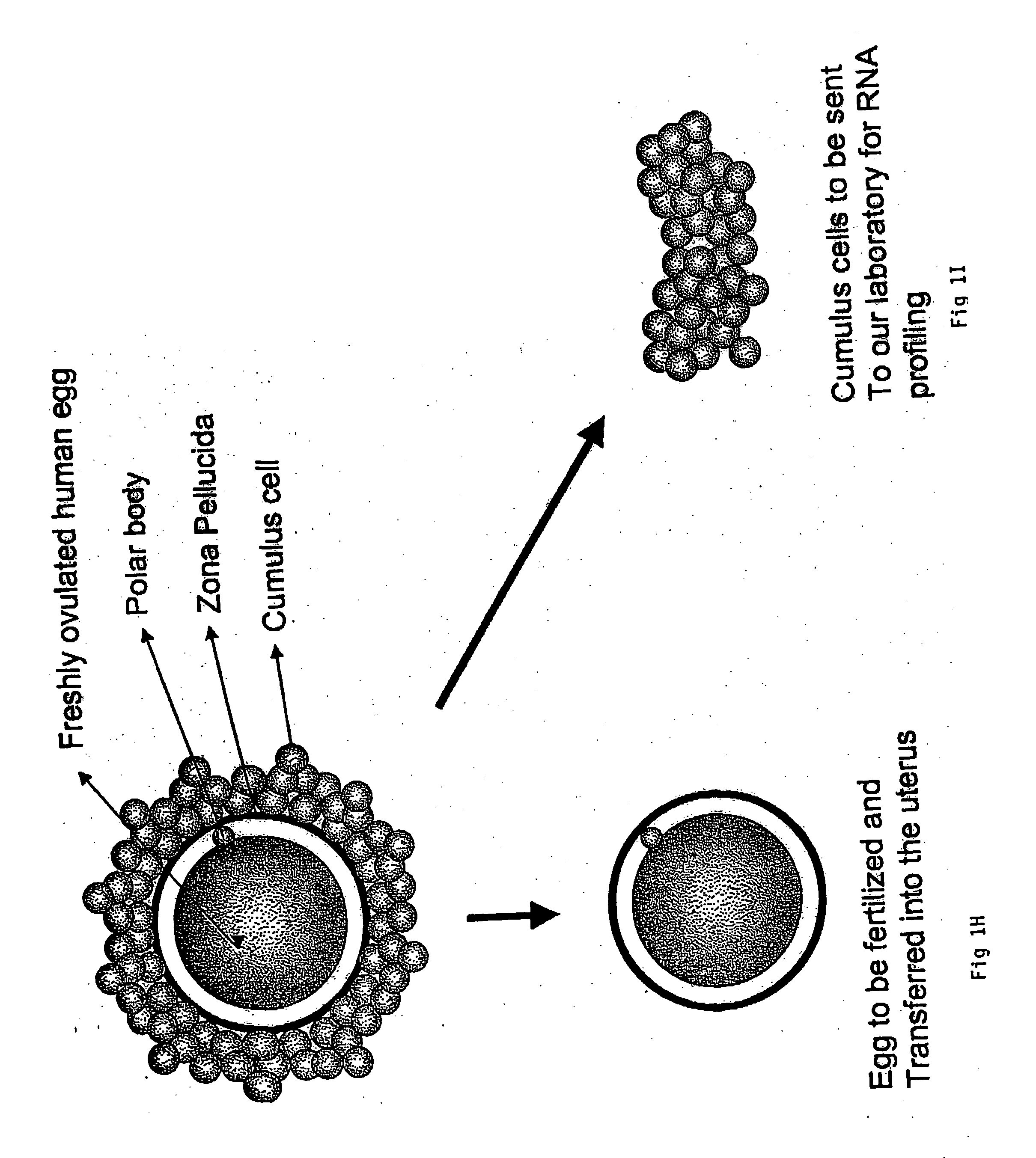
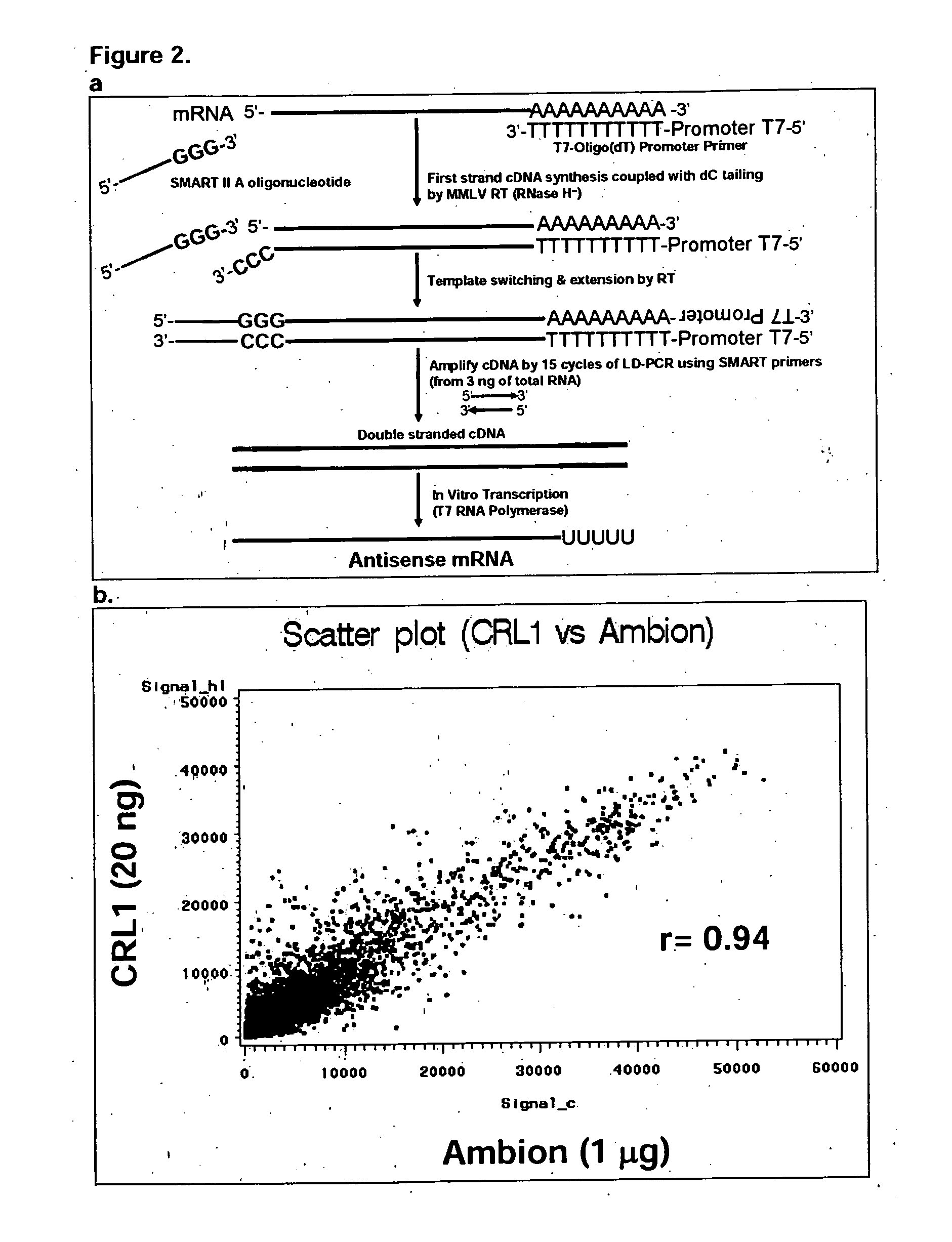
A genetic means of determining whether a female subject produces "pregnancy competent" oocytes is provided. The means comprises detecting the level of expression of one or more genes that are expressed at characteristic levels (upregulated or downregulated) in cumulus cells derived from pregnancy competent oocytes. This characteristic gene expression level, or pattern referred to herein as the "pregnancy signature", also can be used to identify subjects with underlying conditions that impair or prevent the development of a viable pregnancy, e.g., pre-menopausal condition, other hormonal dysfunction, ovarian dysfunction, ovarian cyst, cancer or other cell proliferation disorder, autoimmune disease and the like. Microarrays containing "pregnancy signature" genes or corresponding polypeptides provide another preferred aspect of the invention. Still further, the subject invention can be used to derive animal models, e.g., non-human primate animal models, for the evaluation of the efficacy of putative female fertility treatments. Additionally, an improved RNA amplification protocol is provided herein referred to as the CRL amplification protocol which is suitable for reproducibly amplifying all the RNAs expressed by a cell sample, even when only a few cells are available.
Owner:MICHIGAN STATE UNIV
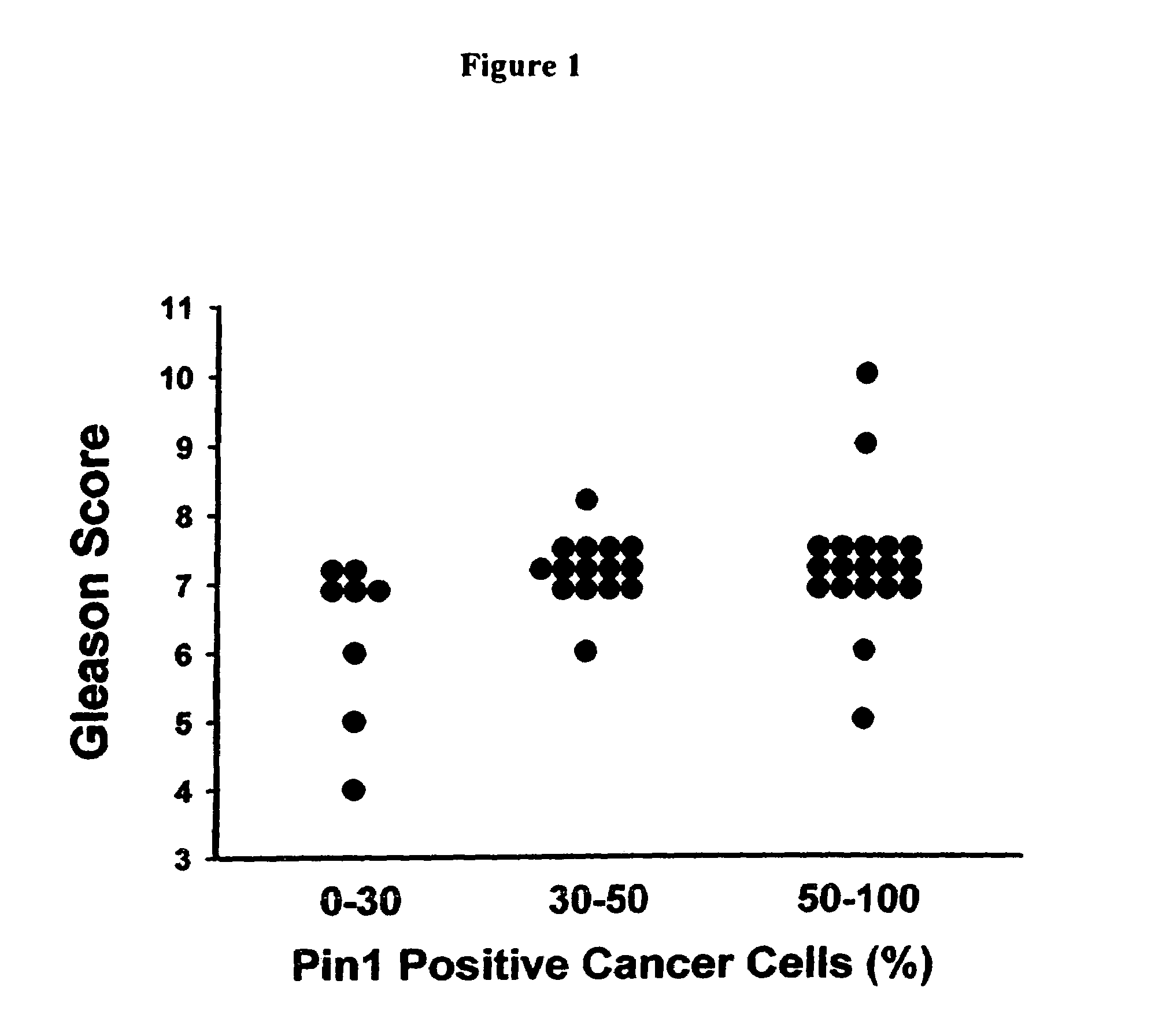
Pin 1 as a marker for prostate cancer
InactiveUS7592145B2Easy diagnosisAvoid failureMicrobiological testing/measurementDepsipeptidesPsa testGrade system
Owner:PINTEON THERAPEUTICS
Methods for PCR and HLA typing using unpurified samples
Provided are methods for amplifying a gene or RNA or sets thereof of interest using a tandem PCR process. The primers in the first PCR or set of PCR reactions are locus-specific. The primers in the second PCR or set of PCR reactions are specific for a sub-sequence of the locus-specific primers and completely consumed during the second PCR amplification. For RNA amplification, the first PCR is reverse transcription and the resulting cDNA(s) provide a template for cRNA synthesis, endpoint PCR or real time PCR. Also provided is a tandem PCR method which accepts raw, completely unpurified mouthwash, cheek swabs and ORAGENE™-stabilized saliva as the sample input, the resulting amplicons serving as the substrate for complex, microarray-based genetic testing. Also provided is a method of allelotyping a gene or set thereof by amplifying the gene(s) using tandem PCR on DNA or RNA comprising the sample.
Owner:GENOMICS USA
Application of Bst DNA polymerase in RNA amplification
InactiveCN105176971AEasy to operateShort reaction timeTransferasesDNA preparationDNA polymerase nuTranscriptase activity
The invention discloses an application of Bst DNA polymerase in RNA amplification. The Bst DNA polymerase has reverse transcriptase activity, and thus RNA is subjected to inverse transcription into cDNA under action of the Bst DNA polymerase. A new RNA amplification detection method is established on the basis of the new discovery that the Bst DNA polymerase has reverse transcriptase activity. The method integrates inverse transcription and subsequent nucleic acid amplification, temperature adjustment is not needed during the whole process, addition of extra reverse transcriptases is not needed, operation is simple, reaction time is shortened, and new methodology reference and technical support are provided for RNA inverse transcription and amplification detection. The discovered Bst DNA polymerase can perform an inverse transcription reaction at a high temperature, which facilitates to open and combine RNAs with complex secondary structures, and therefore the problem is solved that reverse transcriptases of AMV or the like are difficult to amplify RNA advanced structure areas.
Owner:QINGDAO NAVID BIOTECH CO LTD
Method for selectively blocking hemoglobin RNA amplification
InactiveUS20060234277A1Growth inhibitionSugar derivativesMicrobiological testing/measurementWhole blood unitsWhole blood sample
This invention provides oligonucleotides, compositions, kits, and methods for specifically blocking amplification of hemoglobin mRNA in a sample of RNA. The oligonucleotides and methods are particularly advantageous for analyzing samples of RNA extracted from whole blood samples.
Owner:AMGEN INC
Reagent for nucleic acid real-time isothermal amplification, amplification system and amplification method and application of amplification system
InactiveCN108642145ARealize quantitative detectionAchieve hydrolysisMicrobiological testing/measurementMicroorganism based processesFluorescenceLoop-mediated isothermal amplification
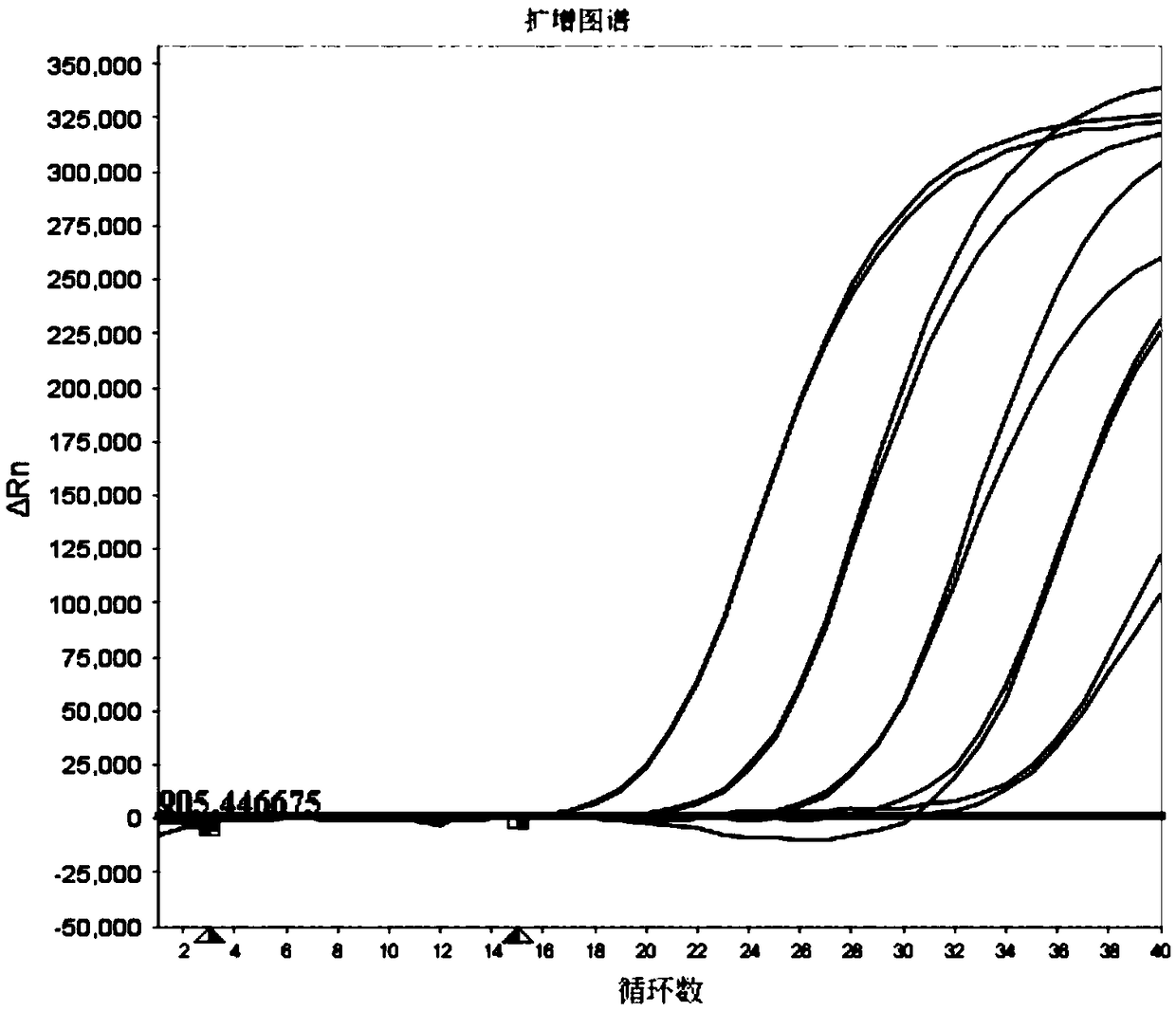
The invention relates to a reagent for nucleic acid isothermal amplification. The reagent comprises RNA polymerase, DNA polymerase with reverse transcription activity and RNase H, wherein the DNA polymerase with reverse transcription activity has 5'-3' DNA polymerization activity and reverse transcription activity and 5'-3' exonuclease activity. The reagent further comprises primers and a fluorescence probe, wherein the primers comprise the primer with an RNA promoter recognition sequence and the second cDNA synthesis primer, and the fluorescent probe is selected from a TaqMan probe and a Molecular Beacon probe. The invention further relates to a nucleic acid isothermal amplification system comprising the reagent and an amplification method thereof, and a kit comprising the nucleic acid isothermal amplification system and application thereof. By using the RNA polymerase, the DNA polymerase with reverse transcription activity and the RNase H, controllable RNA and DNA isothermal amplification is realized; the isothermal amplification is combined with the fluorescent probe technology, which can realize real-time fluorescence detection in the target RNA amplification process; besides,the whole process is completed in the same reaction system, and quantitative detection of nucleic acid is realized. The reagent, the amplification system and the kit are suitable for clinical promotion and application.
Owner:上海默礼生物医药科技有限公司
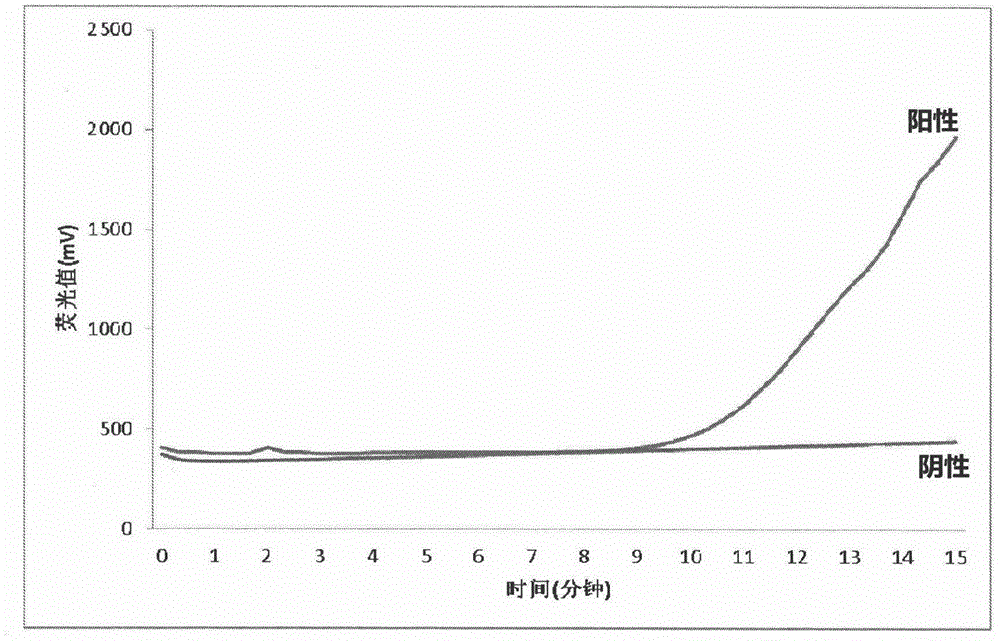
Fluorescent isothermal nucleic acid amplification method
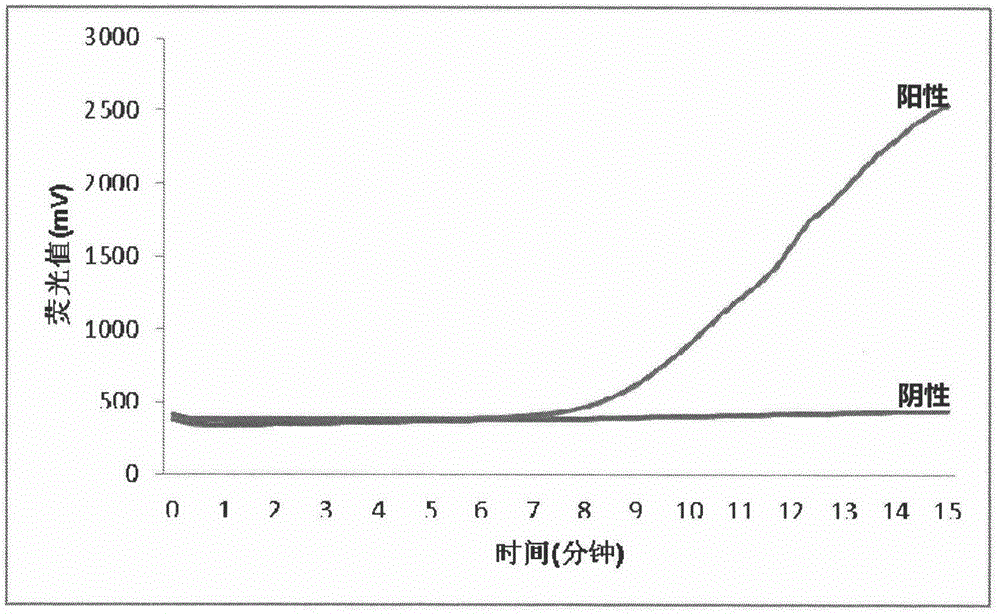
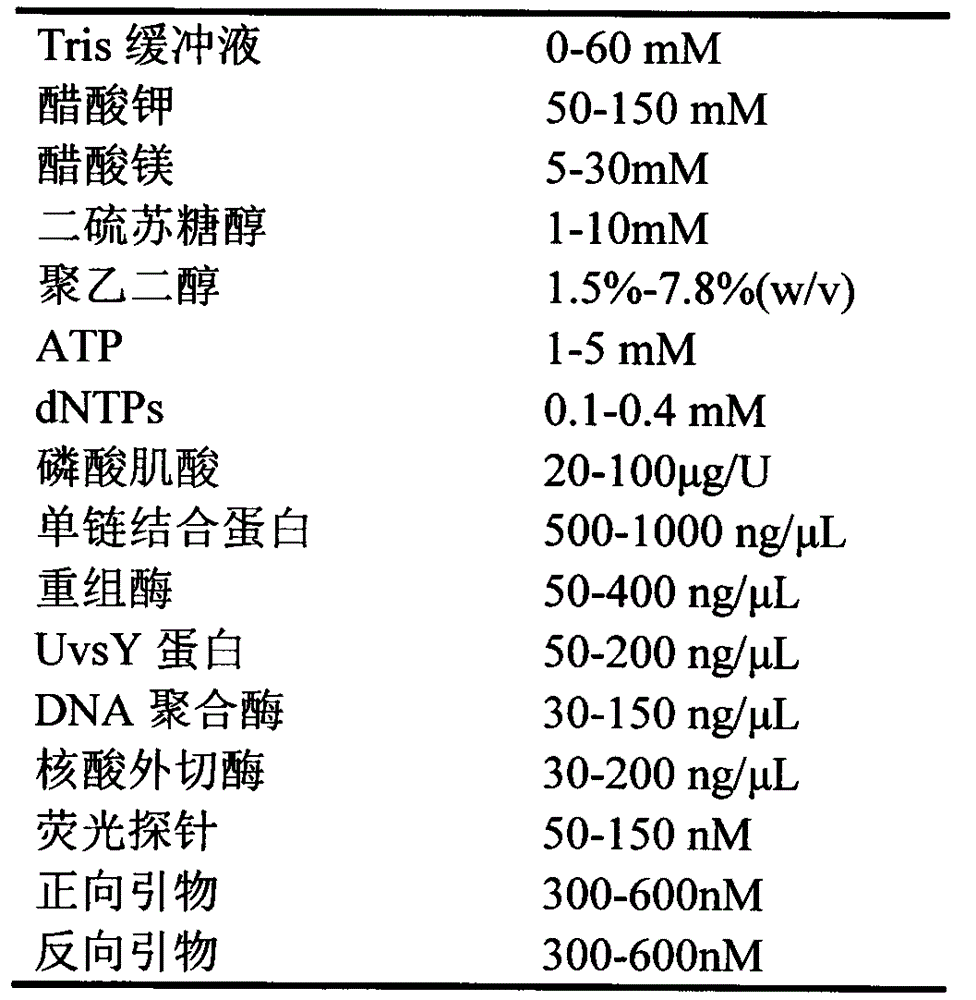
InactiveCN105803045AReal-time monitoring of amplification resultsStrong specificityMicrobiological testing/measurementPhosphateFluorescence
The invention discloses a fluorescent isothermal nucleic acid amplification method. The amplification method comprises the following ingredients: a Tris buffer, potassium acetate, magnesium acetate, dithiothreitol, polyethylene glycol, ATP, dNTPs, creatine phosphate, a single-chain binding protein, a recombinant enzyme, a UvsY protein, a DNA polymerase, an exonuclease, a fluorescent probe and a primer group. The method can be applied to carry out real-time fluorescence monitoring on DNA; and based on the method, addition of an RNA retrovirus cane realize RNA amplification. The isothermal nucleic acid amplification method comprises the steps as follows: (1) extracting DNA or RNA; (2) adding the DNA or RNA to a nucleic acid amplification system; and (3) mixing evenly, adding the mixture into a fluorescence detection instrument, reacting at 35-45 DEG C for 5 to 30 minutes to observe a fluorescence signal. The method has the advanategs of simple operation, fast response, low requirements for operators, and broader range of applications.
Owner:JIANGSU QITIAN GENE BIOTECHNOLOGY CO LTD
Methods for PCR and HLA typing using raw blood
Provided are methods for amplifying a gene or RNA or sets thereof of interest using a tandem PCR process. The primers in the first PCR or set of PCR reactions are locus-specific. The primers in the second PCR or set of PCR reactions are specific for a sub-sequence of the locus-specific primers and completely consumed during the secondary PCR amplification. For RNA amplification, the first PCR is reverse transcription and the resulting cDNA(s) provide a template for cRNA synthesis, endpoint PCR or real time PCR. Also provided is a method of allelotyping a gene or set thereof by amplifying the gene(s) using tandem PCR on DNA or RNA comprising the sample, hybridizing the resulting amplicon or sets thereof to probes with sequences of gene-associated allele variations. A detectable signal indicating hybridization corresponds to an allelotype of the gene or a set of allelotypes for the set of genes.
Owner:GENOMICS USA
Methods and compositions for RNA amplification and detection using an RNA-dependent RNA-polymerase
Methods and compositions are provided for amplifying RNA from an RNA template. Methods and compositions are further provided for detecting an RNA in an RNA containing sample. Compositions used for the amplification of RNA from RNA include a RNA-dependent RNA-polymerase, a RNA helicase, and an energy source. Illustrative RNA-dependent RNA-polymerase enzymes are derived from the tomato or tobacco plant, while illustrative RNA helicase enzymes include the eIF4A and eIF4B proteins.
Owner:QIAGEN NORTH AMERICAN HLDG INC
Low cycle amplification of RNA molecules
InactiveUS20060057622A1Reduce RNA degradationMicrobiological testing/measurementFermentationGeneticsDistortion
The compositions and methods afford an almost unlimited linear RNA amplification from a few cells with minimal differences in the relative abundance of amplified RNAs and their parent mRNA (sample distortion).
Owner:UNIV OF FLORIDA
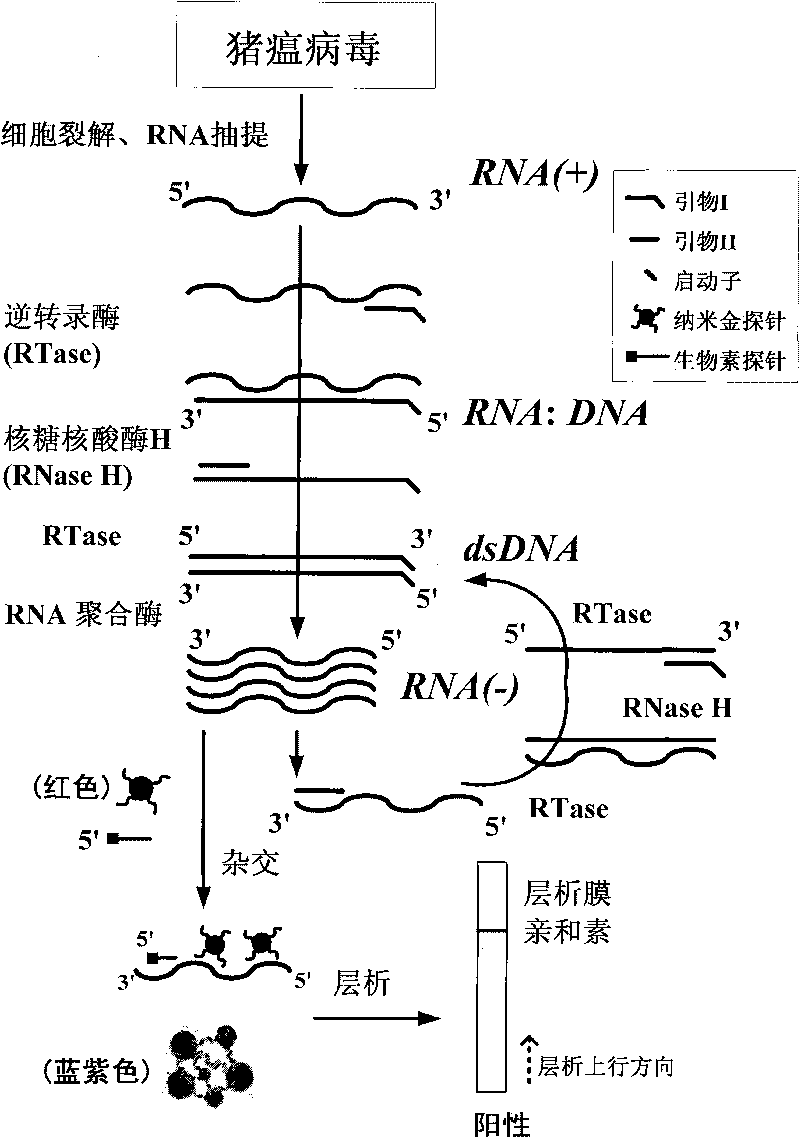
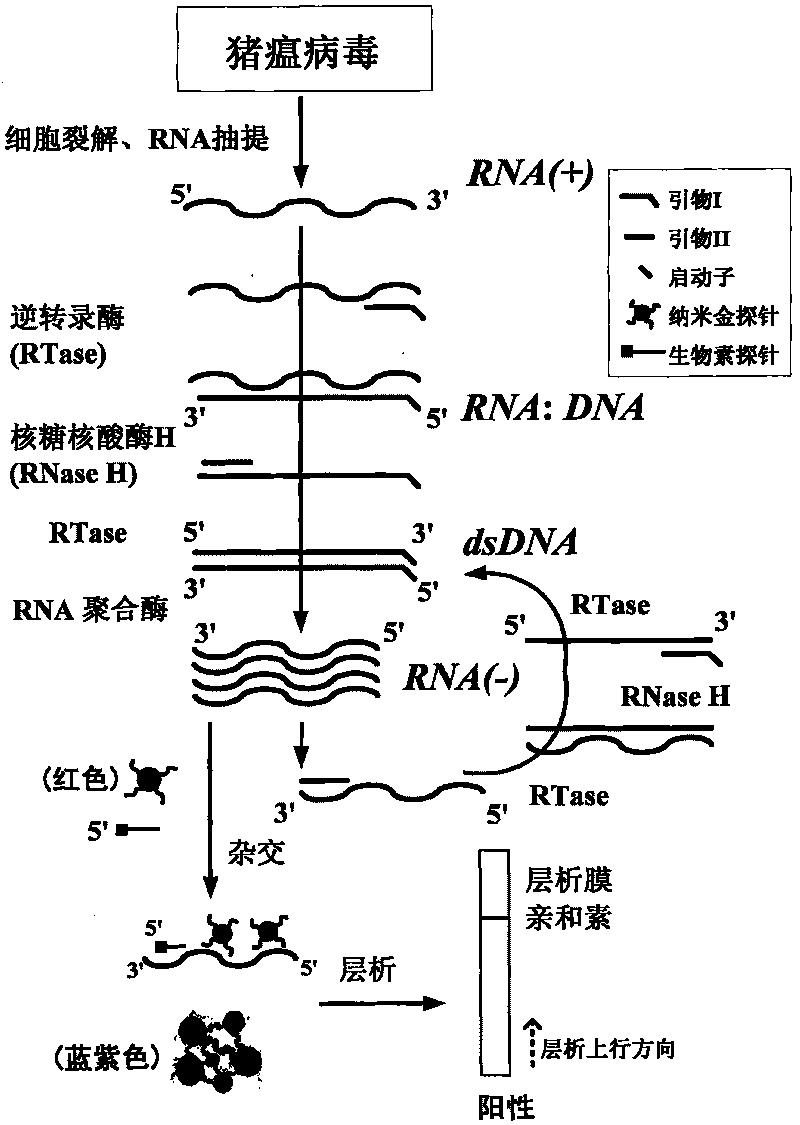
Classical swine fever virus detecting method and kit thereof by nucleic acid isothermal amplication
InactiveCN101760563AEasy and intuitive results observationAvoid False Positive Test ResultsMicrobiological testing/measurementChemiluminescene/bioluminescenceReverse transcriptaseChromogenic
The present invention relates to a classical swine fever virus (CSFV) detecting method and kit thereof by nucleic acid isothermal amplication. The detection includes the steps in sequence of unwinding the RNA template of CSFV under 60 to 70 DEG C and then cooling, adding with reverse transcriptase, ribonuclease H, RNA polymerase, performing the isothermal amplication under 37 to 42 DEG C conducted by primer to obtain RNA amplicon, then detecting the RNA amplicon by gold nanometer probe and capture probe, and obtaining the detecting result through chromatography cross-chromogenic reaction. The RNA is positive if chromogenic stripe appears on the cross film and the RNA is negative or else. The kit of the method can detect the RNA of CSFV quickly and accurately.
Owner:SHANGHAI FOSUN PHARMA (GROUP) CO LTD +1
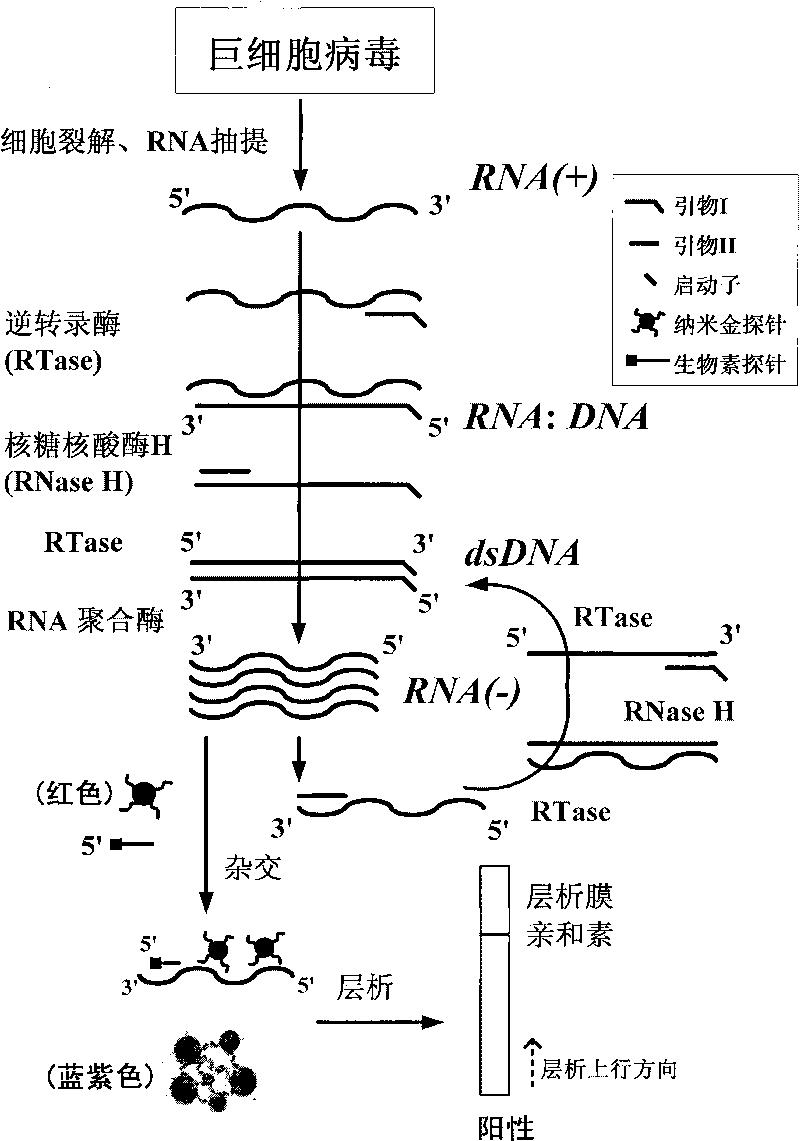
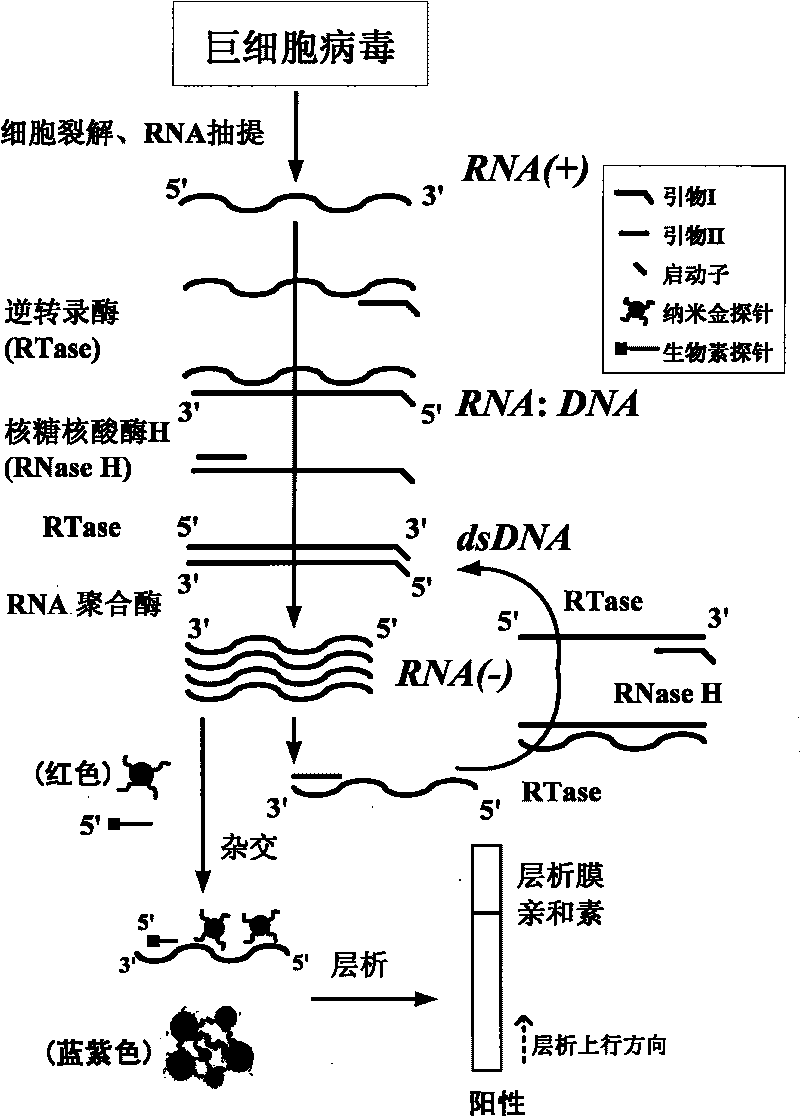
Cytomegalovirus detecting method and kit thereof by nucleic acid isothermal amplication
InactiveCN101760564AAvoid the influence of residual DNAEasy and intuitive to observeMicrobiological testing/measurementChemiluminescene/bioluminescenceReverse transcriptaseChromogenic
The present invention relates to a cytomegalovirus (CMV) detecting method and a kit thereof by nucleic acid isothermal amplication. The detection includes the steps in sequence of unwinding the mRNA template of CMV under 60 to 70 DEG C and then cooling, adding with reverse transcriptase, ribonuclease H, RNA polymerase, performing the isothermal amplication under 37 to 42 DEG C conducted by primer to obtain RNA amplicon, then detecting the RNA amplicon by gold nanometer probe and capture probe, and obtain the detecting result through chromatography cross-chromogenic reaction. The mRNA is positive if chromogenic stripe appears on the cross film and the RNA is negative or else. The kit of the method can detect the mRNA of CMV quickly and accurately.
Owner:SHANGHAI FOSUN PHARMA (GROUP) CO LTD +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com