Primer for amplifying short-chain RNA (ribonucleic acid) and related method thereof
A short-chain, reverse primer technology, applied in biochemical equipment and methods, DNA/RNA fragments, DNA preparation, etc., can solve the problems of no relevant literature, etc., and achieve the effect of reducing synthesis cost, simple design, and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1: Stem-loop RT primer combined with probe for quantitative detection of hsa-miR-30d by real-time PCR
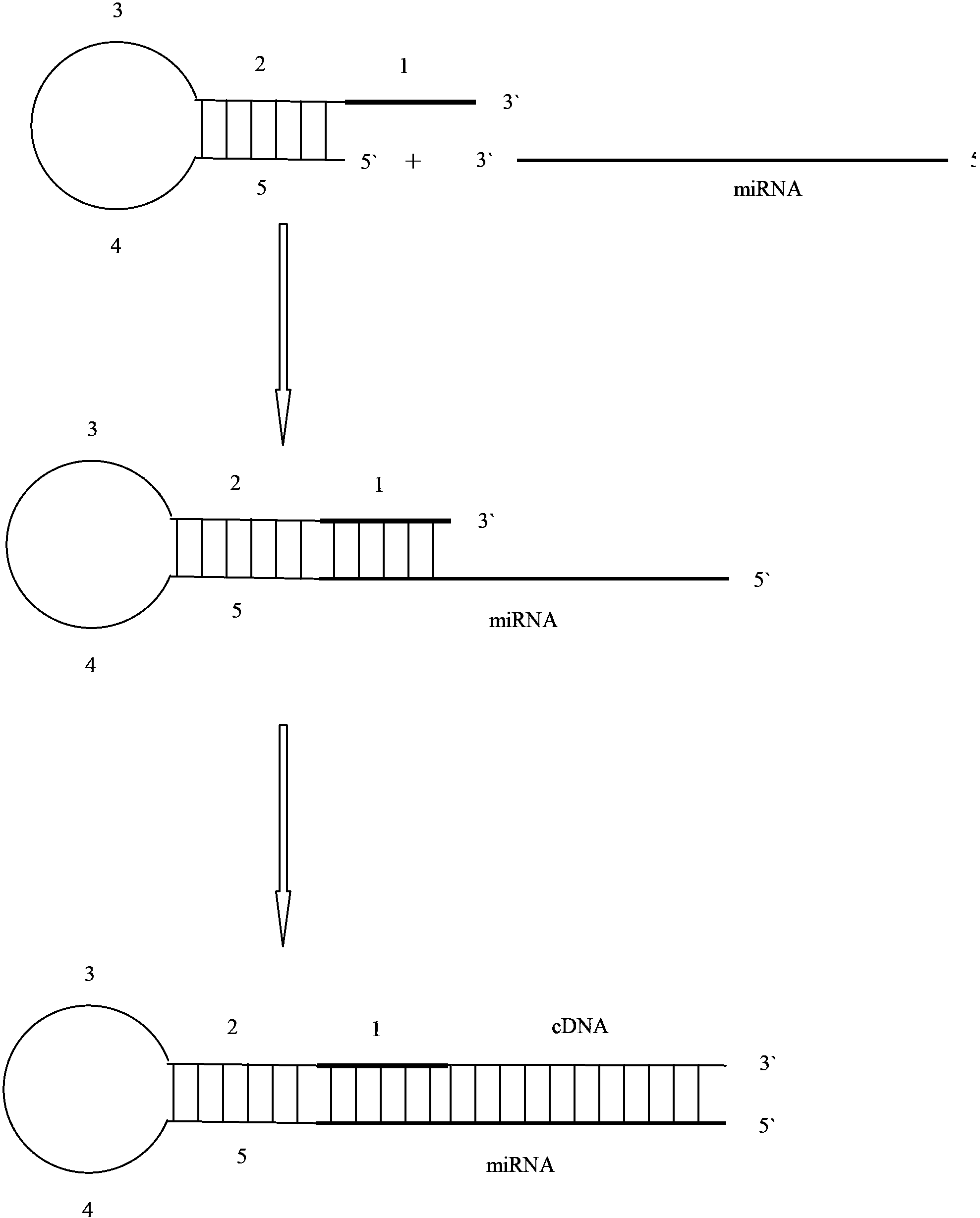
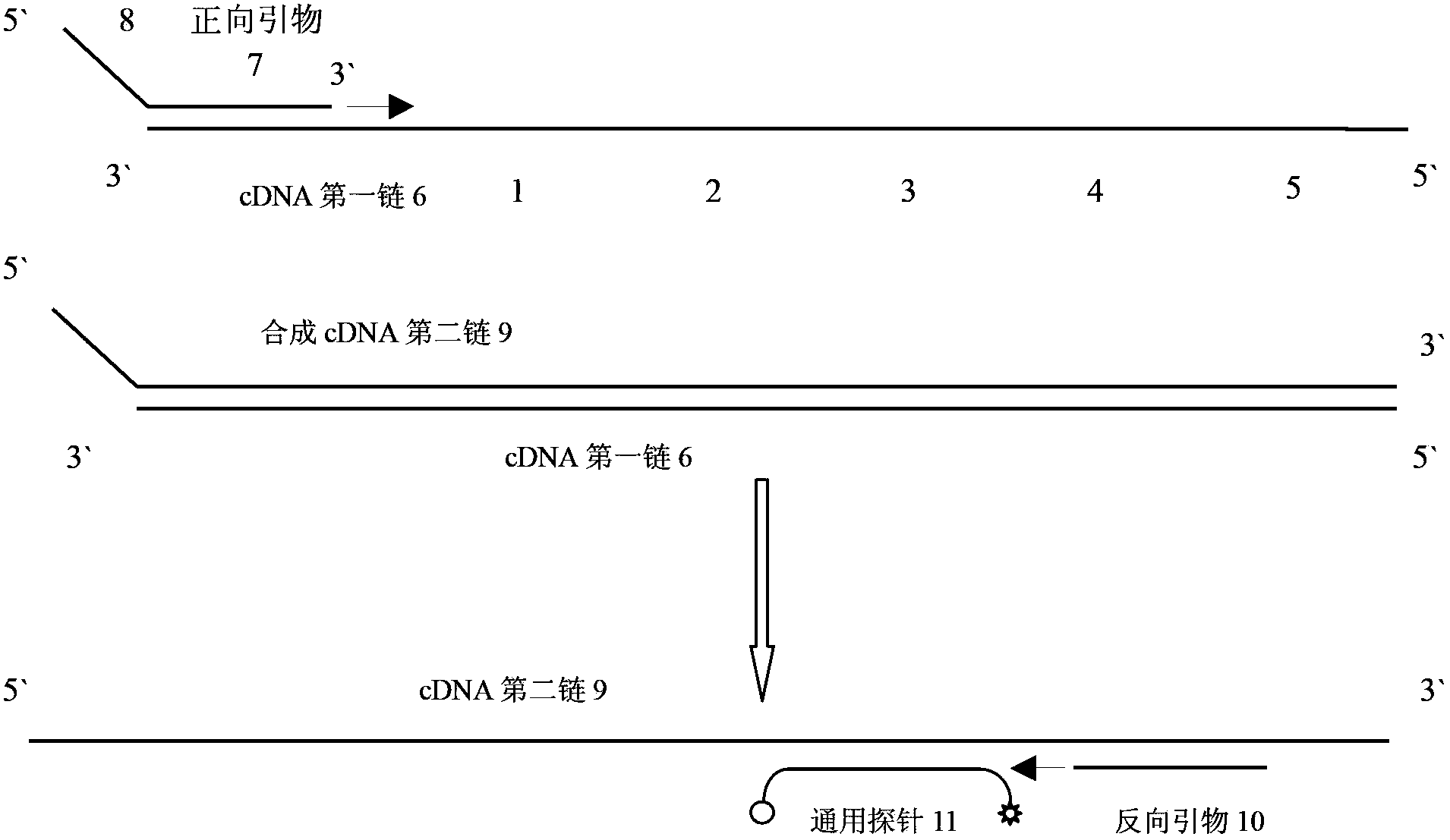
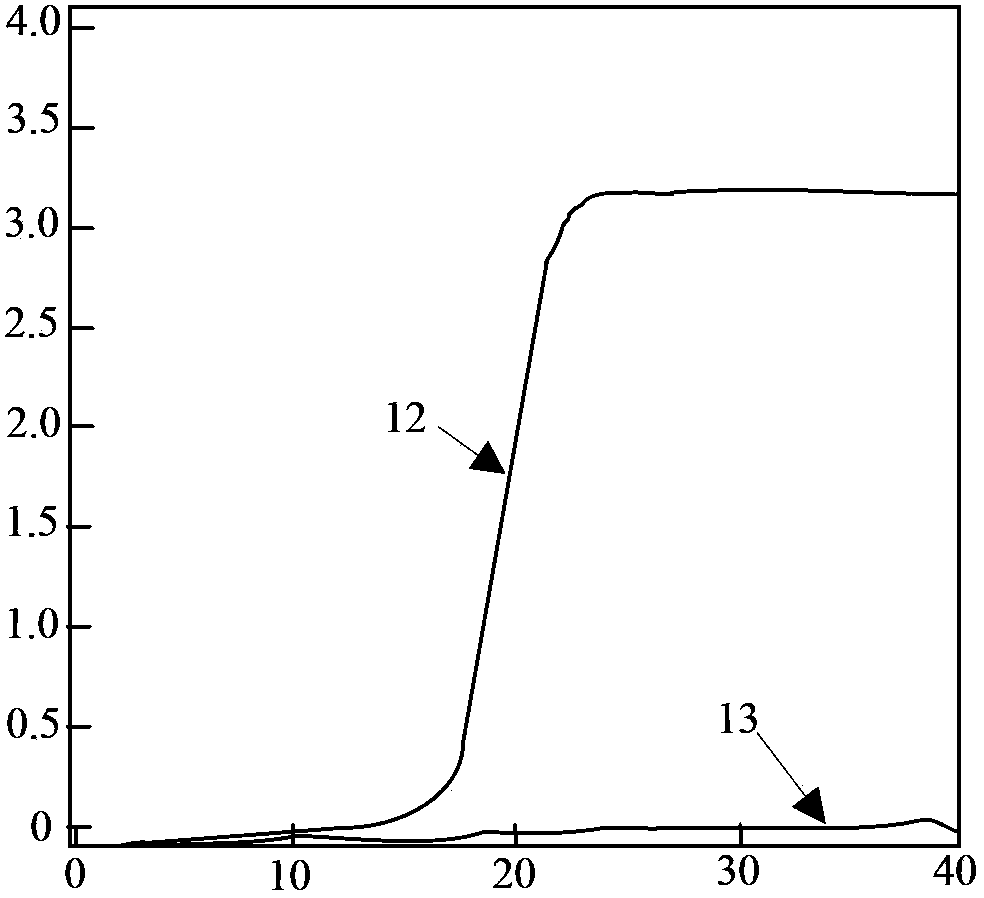
[0045] In order to investigate the specificity and effectiveness of stem-loop primers in real-time PCR detection, the PCR templates were the precursor hsa-mir-30d
[0046] (5`GUUGU UGUAAACAUCCCCGACUGGAAG CUGUAAGACACAGCUAAGCUUUCAGUCAGAUGUUUGCUGCUAC 3`underlined base is the sequence of the mature body) and mature hsa-miR-30d (5`UGUAAACAUCCCCGACUGGAAG3`) standard RNA samples, and a series of diluted hsa-miR-30d standard cDNA samples. The total volume of the RT reaction system is 15 μl, including 1.5 μl 10× buffer, 50 U / μl reverse transcriptase 1.0 μl, 100 mM dNTP 0.15 μl, 20 U / μl RNA inhibitor 0.19 μl, 1 μM RT primer 3 μl, 0.01 μM template RNA 5 μl, DEPC water 4.16 μl. The reverse transcription PCR reaction was carried out on a BIO-RAD MyCycler qualitative PCR instrument. The reaction conditions are: 16°C for 30min, 42°C for 30min, 85°C for 5min, 4°C∞. The t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com