Single-chain fragment antibody-polypeptide amalgamation protein and uses thereof
A fusion protein and fragment technology, which is applied in the fields of biology and medicine, can solve the problems of weak gene expression effect and cannot meet clinical application, and achieve the effect of improving expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1. Construction of a plasmid with a bidirectional function of binding Her2 receptor and carrying siRNA drugs.
[0039] After finding the full-length sequence of the Her2 single-chain fragment antibody from the gene library, primer software was used to design the relevant primers.
[0040] Designed Her2-ScFv primers:
[0041] Positive primer: NcoI is an endonuclease
[0042] 5’-GGGCCATGGCCCAGGTGCAGCTGTTGCAGTCTGGGGCAGAG-3’
[0043] Reverse primer: NotI is an endonuclease
[0044] 5’-TTGCGGCCGCTCCGGAATTCACCTAGGACGGTCAGCTTGGTCCC-3’
[0045] Designed protamine peptide primers:
[0046] Positive primer: NotI is an endonuclease
[0047] 5’-CCGGAGCGGCCGCAATGGCCAGGTACAGATGCTG-3’
[0048] Reverse primer: PpuM I as an endonuclease, stop codon and 6 histidines
[0049] 5’-GCCGGGTCCCAGGAAAGGATCAGATCTGCATTAATGGTGGTGGTGATGATGAGATCTGTGTCTTCTACATCTCGGTCTG-3’
[0050] The relevant target fragments are cloned by PCR. The PCR reaction system of Her2-ScFv is 1:50ul system:
[0051] ddH 2 O 37ul
[005...
Embodiment 2
[0101] Example 2 Small amount extraction of pACgp67B-Her2-scfv-protamine plasmid
[0102] The bacteria were collected by centrifugation at 12000g×1min. (Generally, 3-4ml bacterial solution is used for one part extraction), and the supernatant is discarded. Add 250ul ice-bath Buffer S1 (containing RnaseA), vortex and shake to fully suspend the bacteria. Add 250ul Buffer S2 and mix gently 6 times. (This process does not exceed 5 minutes) Add 350ul Buffer S3 and mix gently 6 times. Centrifuge at 12000g×10min. Take the supernatant, pass through the DNA preparation tube, and discard the filtrate at 12000g×1min. Add 700ul BufferW1, 12000g×1min to the DNA preparation tube, and discard the filtrate. Add 500ul Buffer W2, 12000g×1min to the DNA preparation tube, and discard the filtrate. Centrifuge again at 12000g×1min. Add 40ul ddH2O (or EB buffer) to the center of the membrane of the DNA preparation tube, and let it stand at room temperature for 1 min (preheating EB buffer at 50°C...
Embodiment 3
[0103] Example 3. Restriction restriction identification of Her2 single-chain fragment antibody and the plasmid pACgp67B-Her2-scfv-protamine constructed by the protamine polypeptide
[0104] Restriction digestion to identify pACgp67B-Her2-scfv-protamine plasmid fragment:
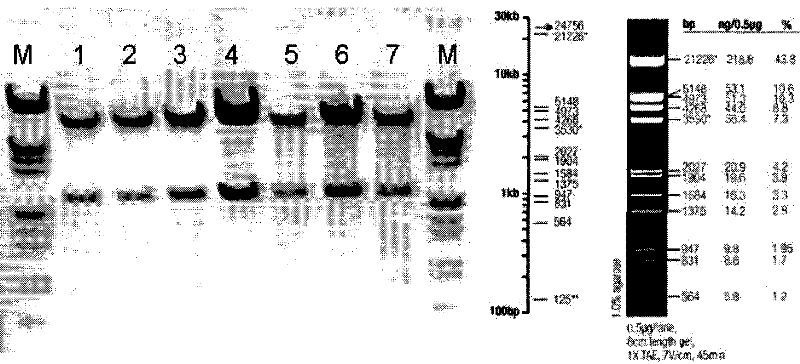
[0105] ①. The restriction site of BamHI (G^GATCC) is at: 4259; the restriction site of XhoI (C^TCGAG) is at: 1902; so the size of the band after restriction is: 2357bp, the position is accurate. Such as figure 1 As shown, pACgp67B-Her2-scfv-protamine was identified by digestion with BamHI and XhoI. M is a marker, 1, 2, 3, 4, 5, 6, 7 are different clones of bacteria amplified and extracted plasmid DNA. After restriction digestion, there is a corresponding band at 2027 bp above, which is consistent with the expected band position.
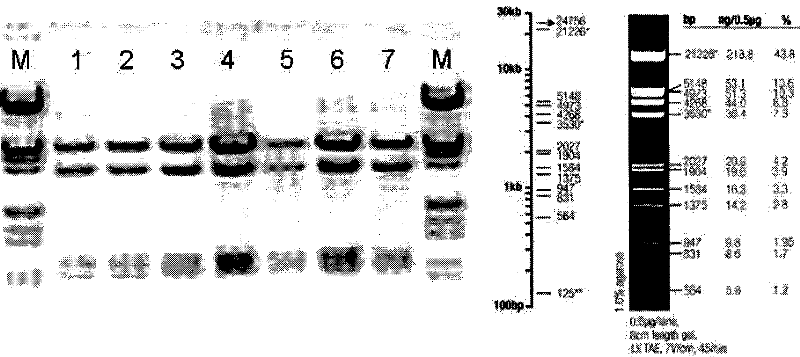
[0106] ②. The restriction site of HindIII (A^AGCTT) is at: 2, 5328, 6256, 7292, so the size of the four bands after restriction is: 5326bp, 3423bp, 1036bp, 928bp, the position is accurat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com