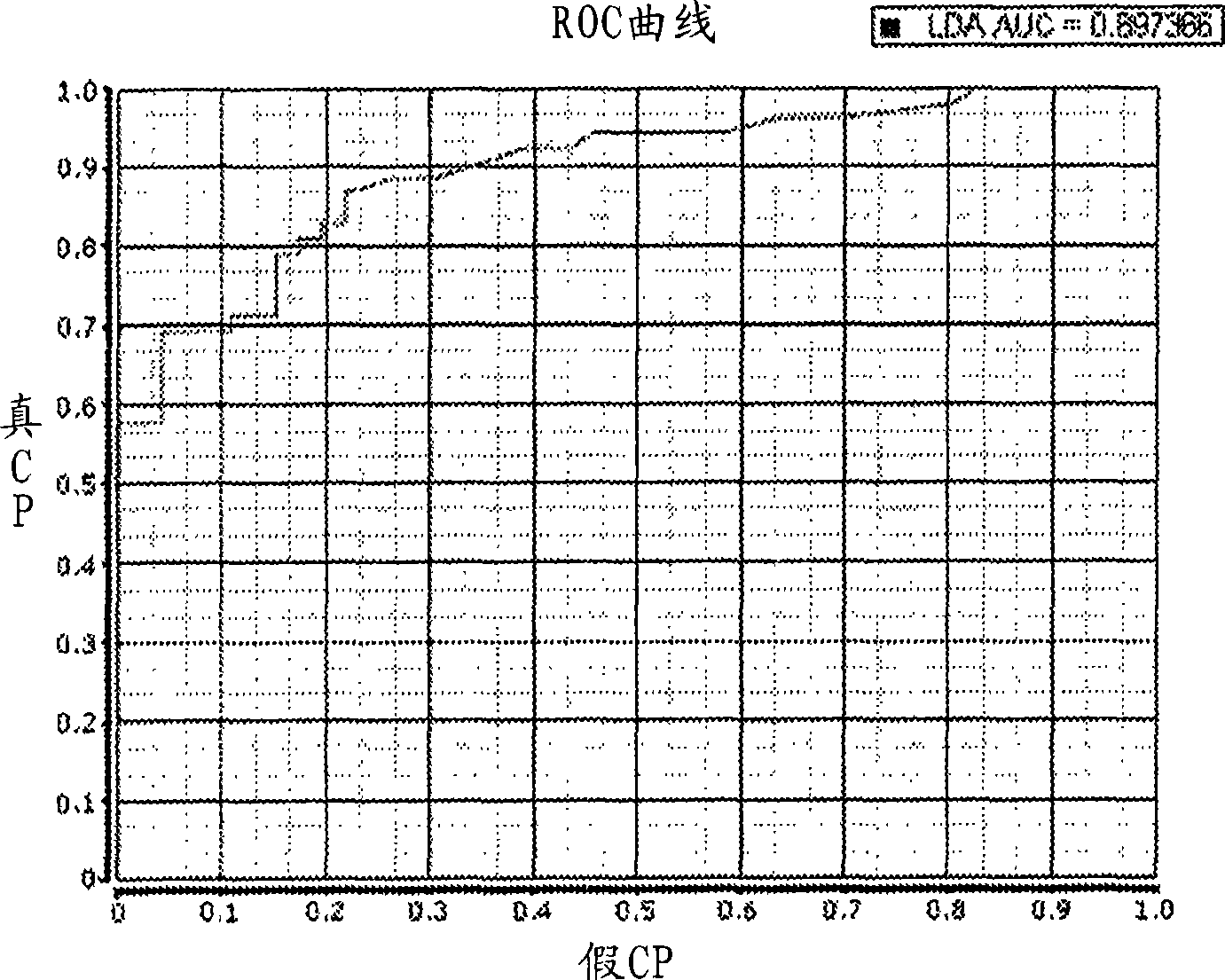
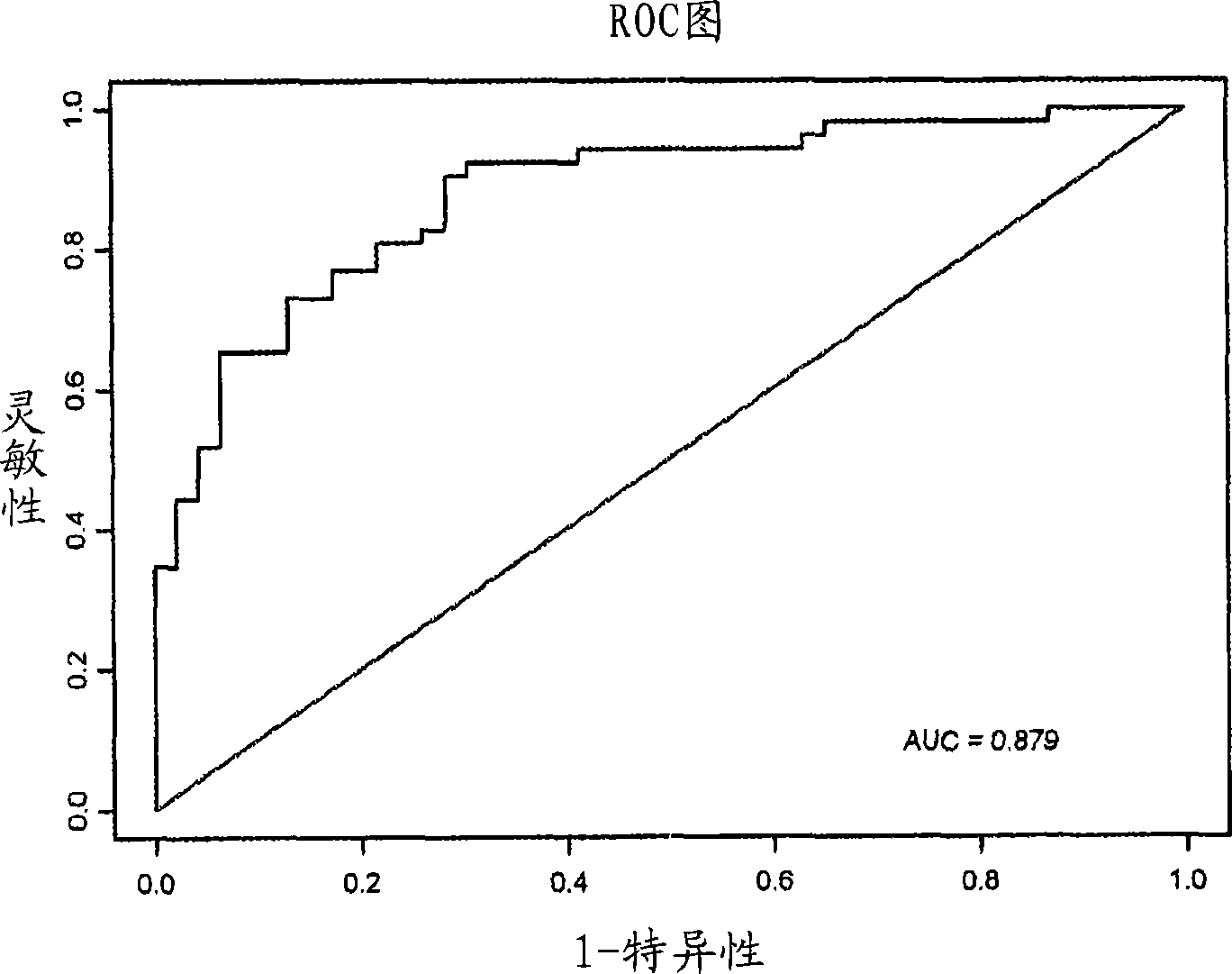
Thyroid fine needle aspiration molecular assay
A technology of thyroid cancer and probe, which is applied in the direction of microbial determination/inspection, analytical materials, measuring devices, etc., and can solve the problems of lack of sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0089] Materials and methods
[0090] tissue sample
[0091] Fresh-frozen samples of benign thyroid disease, follicular adenoma, follicular carcinoma, and papillary carcinoma were obtained from various commercial suppliers, including Genomics Collaborative, Inc. (Cambridge, MA), Asterand (Detroit, MI), and Proteogenex ( Los Angeles, CA). All samples were collected according to Institutional Review Board approved protocols. Patient demographic and pathological information was also collected. Review histopathological features of each sample to confirm diagnosis, estimate sample preservation, and tumor content.
[0092] RNA isolation
[0093] All RNA isolations were performed using standard TriZol protocols. Tissues were homogenized in TriZol reagent (Invitrogen, Carlsbad, CA). Total RNA was isolated from TriZol and precipitated with isopropanol at -20°C. The RNA pellet was washed with 75% ethanol, dissolved in water, and stored at -80°C for future use. RNA integrity was ...
Embodiment 2
[0114] microarray analysis
[0115] Labeled cRNA was prepared and hybridized to the High Density Oligonucleotide Array Hu133A Gene Chip (Affymetrix, Santa Clara, CA) containing a total of 22,000 probe sets. Hybridization was performed according to standard protocols provided by the manufacturer. Arrays were scanned using the Affymetrix protocol and scanner. For subsequent analyses, each probe set was considered an independent gene. The expression value of each gene was calculated using Affymetrix gene chip analysis software MAS 5.0. All chips met the following quality control criteria: percentage of "presence" calls, scaling factors, background levels and noise levels must be within + or -3 standard deviations of the mean. All chips used for subsequent analysis have passed these quality control standards. Sample collection for marker selection and independent validation is summarized in Table 1.
[0116] Table 1. Sample collection for labeled training and validation
[0...
Embodiment 3
[0120] Result marker identification
[0121] A. Gene selection
[0122] A total of 98 samples, including 31 primary papillary thyroid tumors, 21 follicular thyroid carcinomas, 33 follicular adenomas, and 13 benign thyroid tissues, were analyzed using the Affymetrix human U133A gene chip. Five gene selection criteria were applied to the entire data set to obtain a limited number of genes for subsequent gene signature or marker identification:
[0123] 1. Consider genes with at least one "Present Call" in the sample set.
[0124] 2. Exclude genes with more than one "present request" in 12 PBL samples.
[0125] 3. Only genes with chip intensities greater than 200 in all samples were selected.
[0126] 4. Using genes that passed the above 3 criteria, we performed a number of analyzes listed in Table 2 to identify genes that were up- or down-regulated in thyroid tumors.
[0127] 5. Finally, genes with a greater than 1.4-fold change in expression were selected.
[0128] Table 2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com