Regenerated chimeric RNAs as biomarkers enriched in human prostate cancer
A technology for prostate cancer and cancer, applied in the fields of cell biology, pharmacology, and molecular biology, can solve the problem of increasing chimeric RNA without research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0168] Identification of novel regenerative chimeric RNAs in prostate cancer
[0169] To identify chimeric RNAs expressed in prostate cancer, the inventors sequenced the transcriptomes of 20 cancer samples and 10 matched benign samples from prostate cancer patients. This patient did not undergo prostatectomy prior to radical prostatectomy. A Genome Sequencer (Illumina) was used for sequencing of these samples to generate output sequence pairs of 36 nucleotide reads. A total of 30 lanes of a genome sequencer (Illumina) were used to generate ~1.3 billion raw sequence reads. Using stringent filters (see Example 9), the inventors obtained nearly 5 million reads that could be uniquely mapped to the human genome and met our additional bioinformatics criteria.
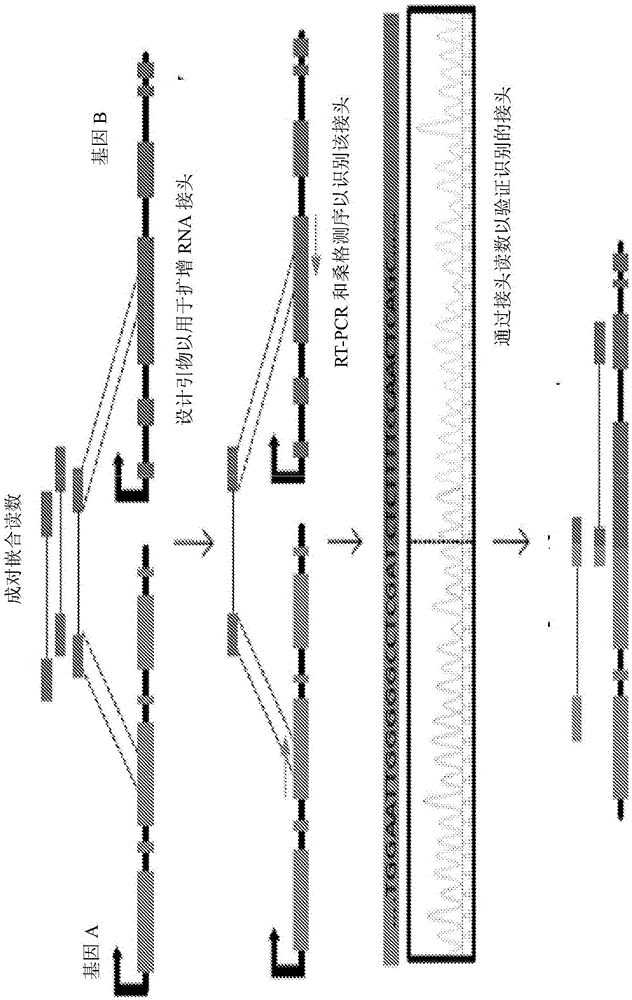
[0170] A strategy for identifying chimeric RNAs is to search for pairs of chimeric reads by each read mapping to a distinct gene in the genome or transcriptome ( figure 1 ). To minimize false positives, the inventors requ...
example 2
[0188] Regeneration of chimeric RNA in cancer vs benign matched samples
[0189] To evaluate its differential expression in cancer vs benign matched samples, the inventors mapped the number of chimeric and splice reads supported by each chimeric RNA and its regeneration rate for each patient. Such as image 3 As shown, most of the chimeric RNAs validated appeared to be highly reproducible in cancer samples, although most of them were also benign matched tissues from radical prostatectomy (however less frequent). This included TMPRSS2-ERG as a control, which is a known cancer specific drug (9). The presence of chimeric RNA in matched benign samples may represent a "field effect" of histologically normal epithelium with multiple premalignant lesions preceding histological changes (17). Additionally, small cancer lesions may be present in some matched benign samples, since the tissue was not histologically evaluated throughout the tissue used for RNA extraction. Nonetheless, s...
example 3
[0196] Expression of Chimeric RNA in Prostate Cancer Cell Lines
[0197] Because the identified chimeric RNAs are easily regenerated in human prostate cancer samples, the inventors believe that these chimeric RNAs can also be present in the established prostate cell lines in some embodiments. Therefore, the inventors tested the expression of these chimeric RNAs in prostate-derived cancer cell lines, including androgen receptor-negative cell lines (PC3 and DU145) and androgen-sensitive cell lines (LNCaP, VCAP, and LAPC4) , and immortalized prostate epithelial cells (PNTla) for comparison with non-cancerous primary human prostate epithelial cells (PREC). RT-PCR results showed that most of the chimeric RNAs among the 32 chimeras in the cancer cell lines (Table 4) showed different expression profiles. However, nine chimeric RNAs were significantly absent in non-cancer PrEC (Table 4 and Figure 4 B). These RNAs included TMEM79-SMG5, the same chimera detected by RT-PCR in cancer ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com