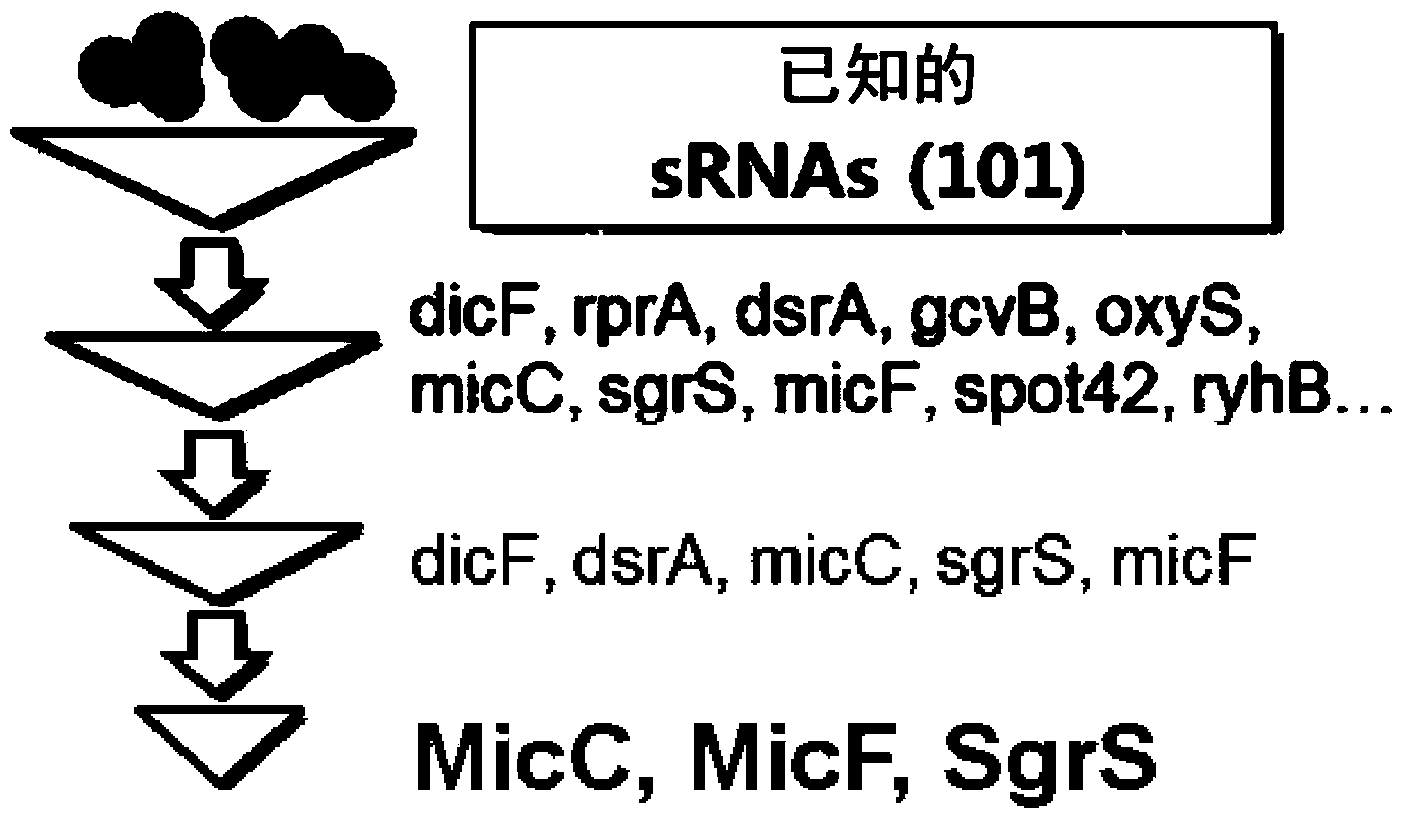
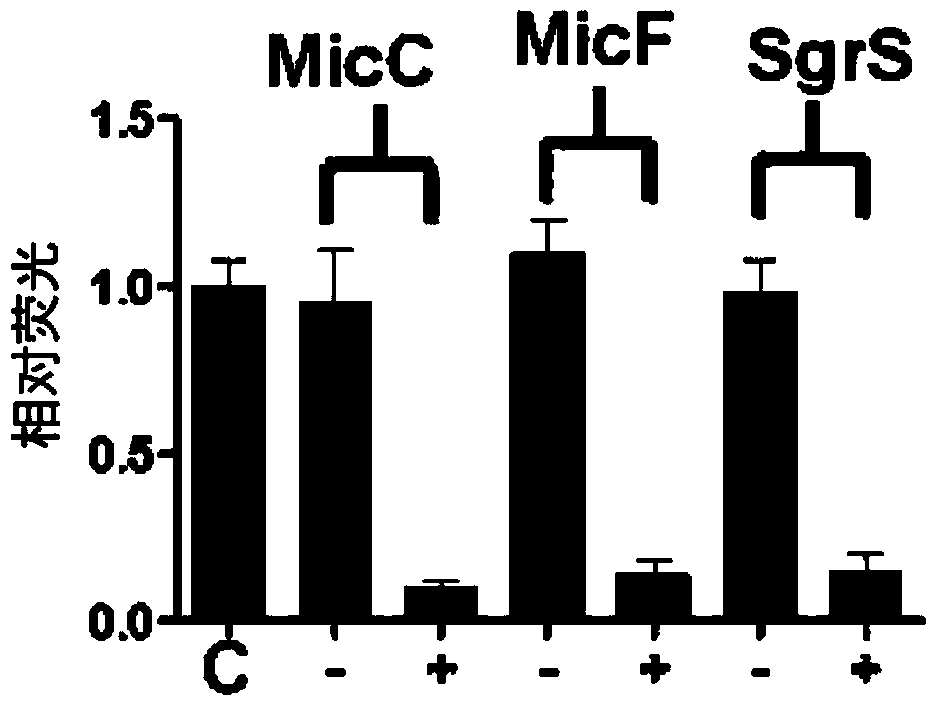
Novel synthesis-regulating srna and method for preparing same
A technology of regulators and transcriptional regulators, applied in the fields of biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., which can solve the problems of difficult gene repair and few or no reports of synthetic sRNA.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0118] Example 1: Performance testing of synthetic sRNA
[0119] 1-1: Construction of synthetic sRNA and construction of pWAS plasmid for regulating expression
[0120] Construct Figure 18 Basic plasmids are shown in , where the expression of synthetic sRNA is suppressed, but can be efficiently expressed if desired. exist Figure 18 In the pWAS plasmid, the target gene mRNA binding region can be inserted into the promoter by site-directed mutagenesis (P R ) and MicC Hfq binding region to construct synthetic sRNA.
[0121] Restriction enzymes used in this and the following examples were purchased from New England Labs (USA), and PCR polymerase was purchased from Invitrogen (USA). Other reagents are indicated separately.
[0122] First, PCR was performed using the pKD46 plasmid (GenBank AY048746.1 (Datsenko et al, PNAS, 97(12): 6640-6645, 2000)) as a template and primers of SEQ ID NO: 2 and SEQ ID NO: 3, and using SacI / EcoRI cloned the araC-PBAD promoter into the pWA pl...
Embodiment 2
[0167] Example 2: Screening of mRNA positions effective for the function of synthetic sRNA
[0168] In order to determine the binding positions of mRNAs effective for the stabilizing function of synthetic sRNAs, synthetic rRNAs were constructed using the pWAS plasmid as a template to bind to various sites of DsRed2.
[0169] In the same manner as described in Example 1-3, the region introduced into each synthetic sRNA and the region into base pairing with DsRed2 mRNA, which was introduced including the Hfq binding site of MicC, were amplified by PCR using the following primers in the pWAS plasmid.
[0170] exist Figure 4 Among them, sequence A utilizes the primer of SEQ ID NO:27 and SEQ ID NO:28 to obtain; Sequence B1 utilizes the primer of SEQ ID NO:33 and SEQ ID NO:34 to obtain; Sequence B2 utilizes the primer of SEQ ID NO: 35 and the primers of SEQ ID NO:36 are obtained; Sequence C1 is obtained by utilizing the primers of SEQ ID NO:37 and SEQ ID NO:38; Sequence C2 is o...
Embodiment 3
[0198] Example 3: Construction and cross-reactivity determination of sRNA for other target mRNAs
[0199] In order to test whether the sRNA according to the present invention can inhibit the expression of mRNAs of genes other than DsRed2, a sequence complementary to the RBS of each mRNA of the LuxR, AraC and KanR genes was inserted into the MicC structure by site-directed mutagenesis in order to bind to the RBS . exist Figure 6 In , the sequence complementary to the RBS of each target gene is underlined and shown in red.
[0200] Synthetic sRNA anti-LuxR was constructed by site-directed mutagenesis using the pWAS plasmid (containing the MicC-based construct) as a template and primers of SEQ ID NO:57 and SEQ ID NO:58, anti-AraC was constructed using SEQ ID NO :59 and the primers of SEQ ID NO:60 were constructed, and anti-KanR was constructed using the primers of SEQ ID NO:61 and SEQ ID NO:62. In order to detect the expression level of each gene, the fusion protein of each...
PUM
| Property | Measurement | Unit |
|---|---|---|
| control rate | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com