4-10 decoder design method based on strand displacement
A design method and decoder technology, applied in the field of DNA4-10 decoders, can solve the problems of complex reaction factors, DNA logic circuits not being designed, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
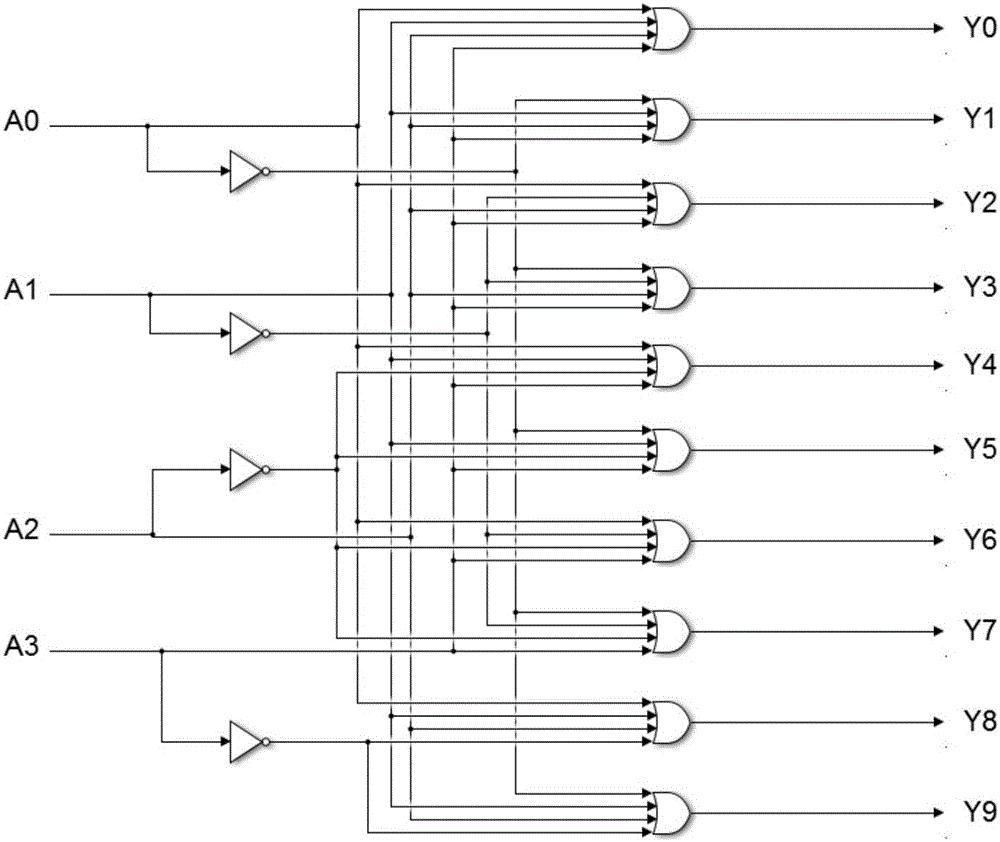
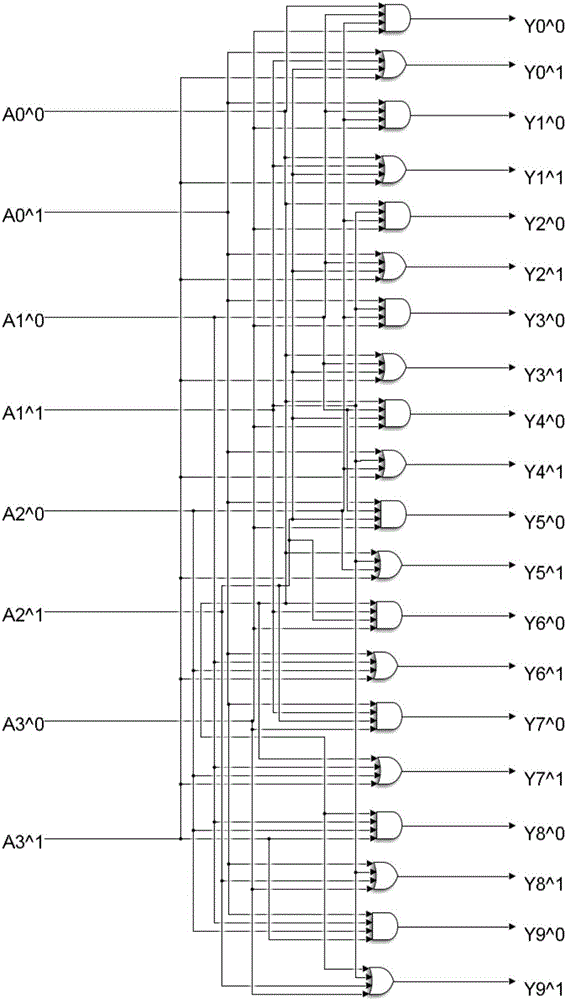
[0034] The embodiments of the present invention are carried out on the premise of the technical solutions of the present invention, and detailed implementation methods and specific operation processes are given, but the protection scope of the present invention is not limited to the following embodiments. Because the threshold concentration coefficient is not fixed and the concentration of the DNA input signal chain is not fixed, the present invention uses the threshold concentration in the fan-out gate module, the AND gate module and the OR gate module as 0.4, 3.2, 0.6 times the unit amount and The high and low concentrations in the input DNA signal chain are respectively set to 0.9 and 0.1 times the unit amount as an example. Design method of the present invention is as follows:
[0035] Step 1: Set the concentration of the threshold gate in the 10 output fan-out modules to 0.4 times the unit volume, set the concentration of the threshold gate in the 4 input AND gate modules...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com