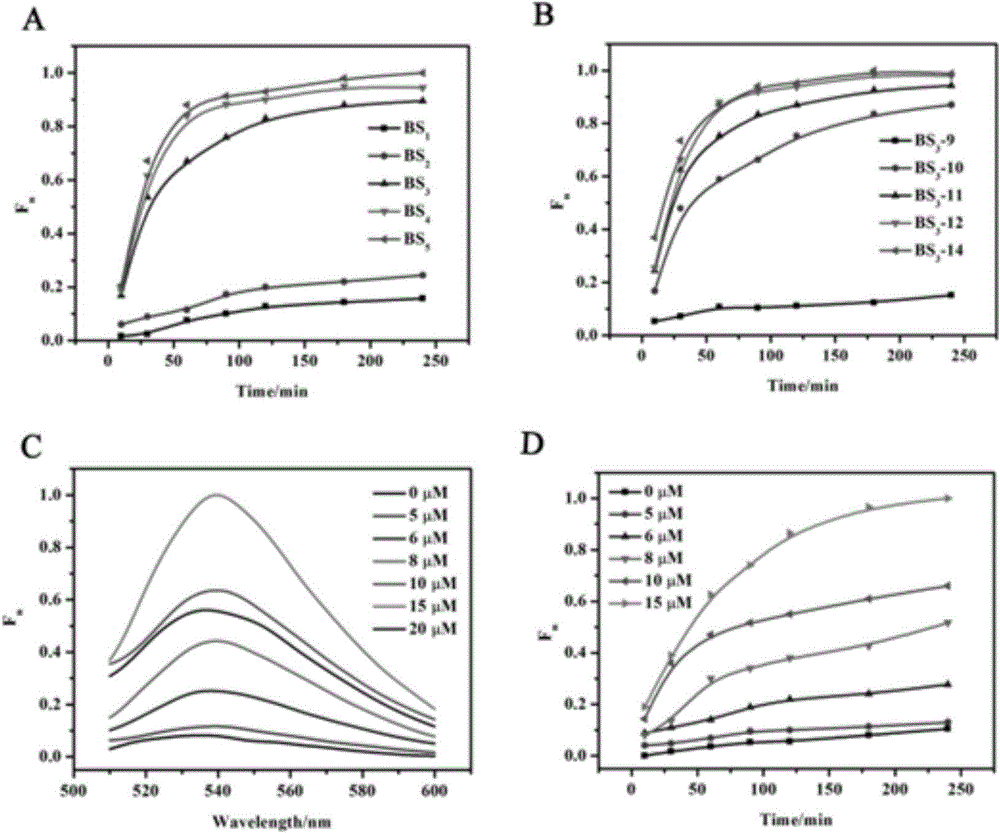
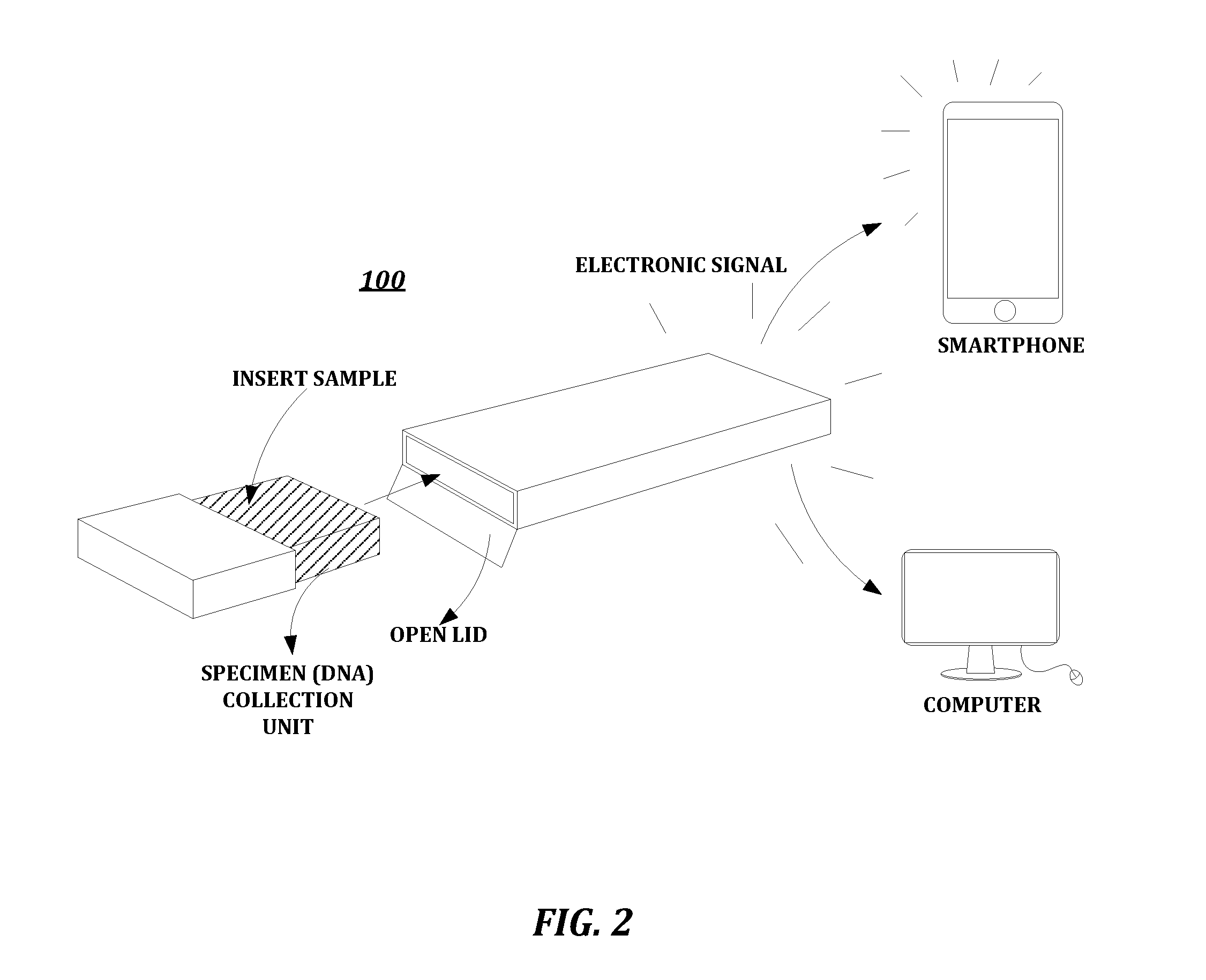
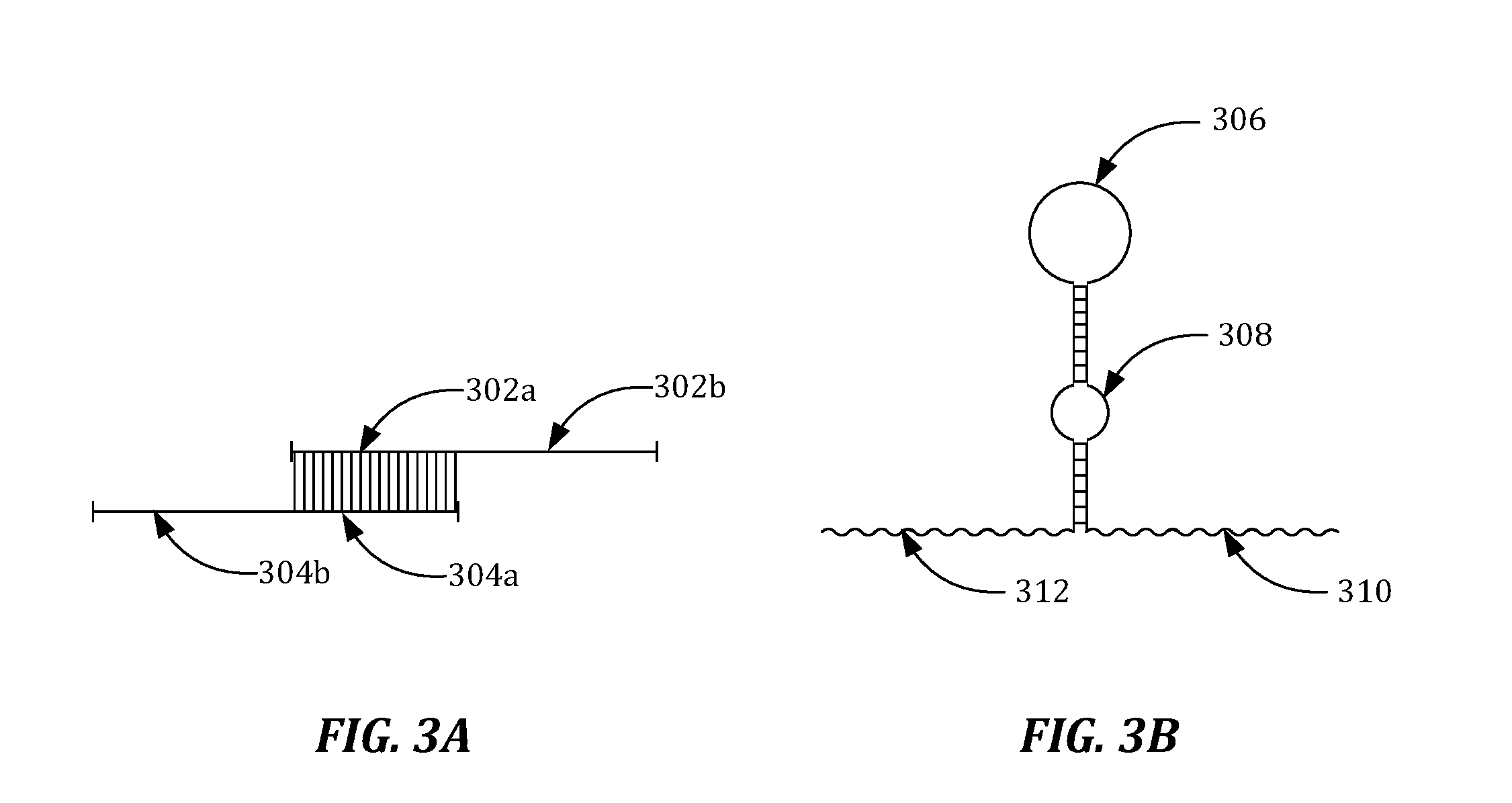
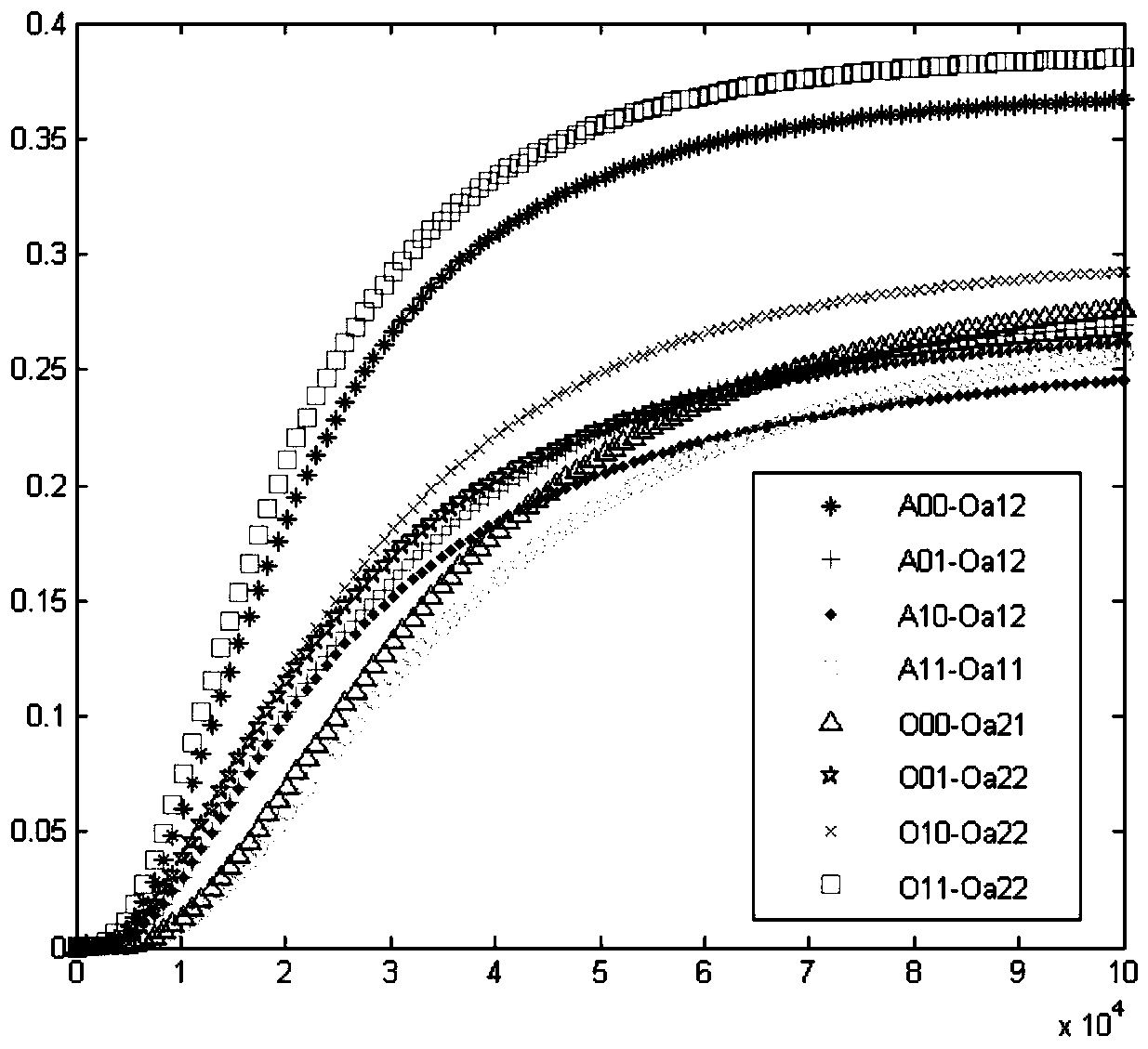
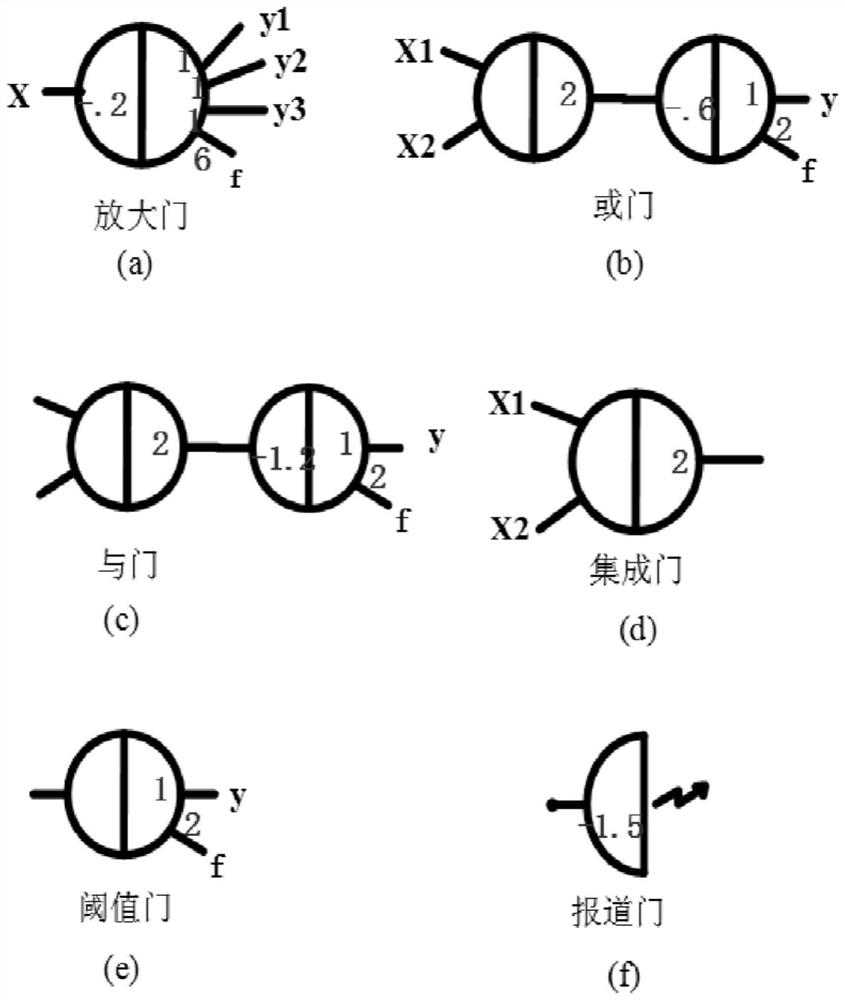
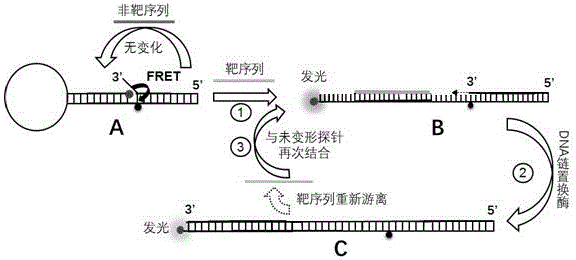
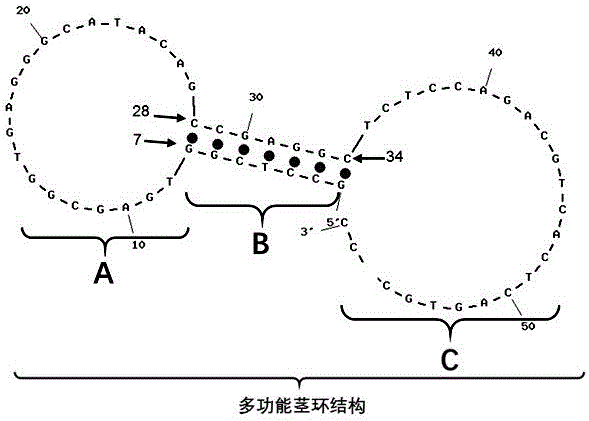
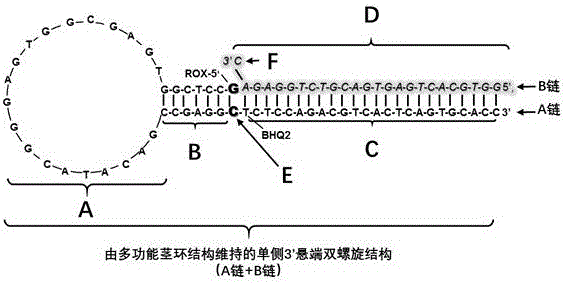
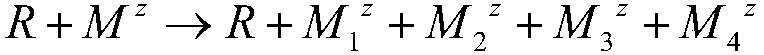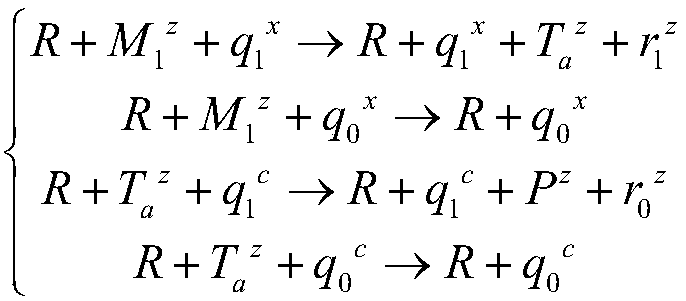Patents
Literature
52 results about "Dna strand displacement" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
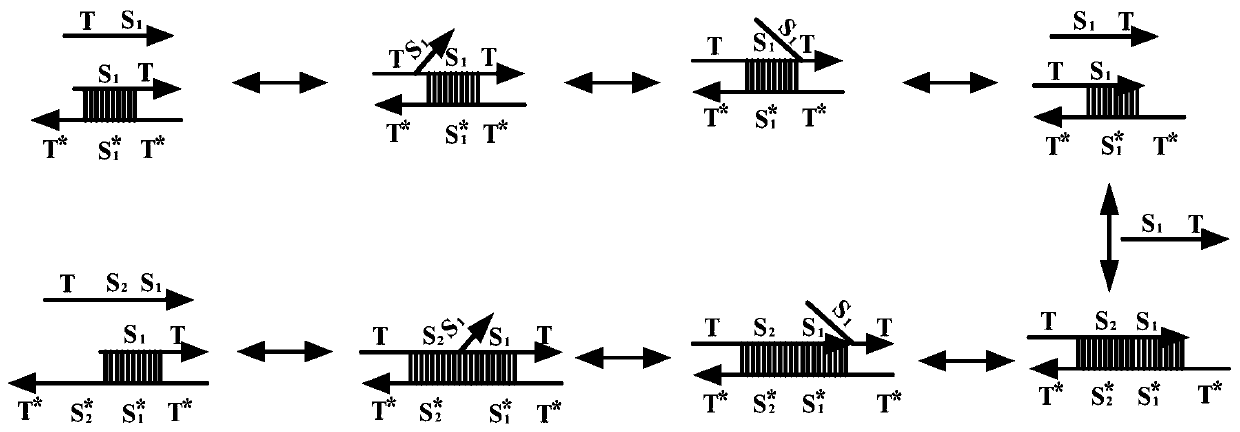
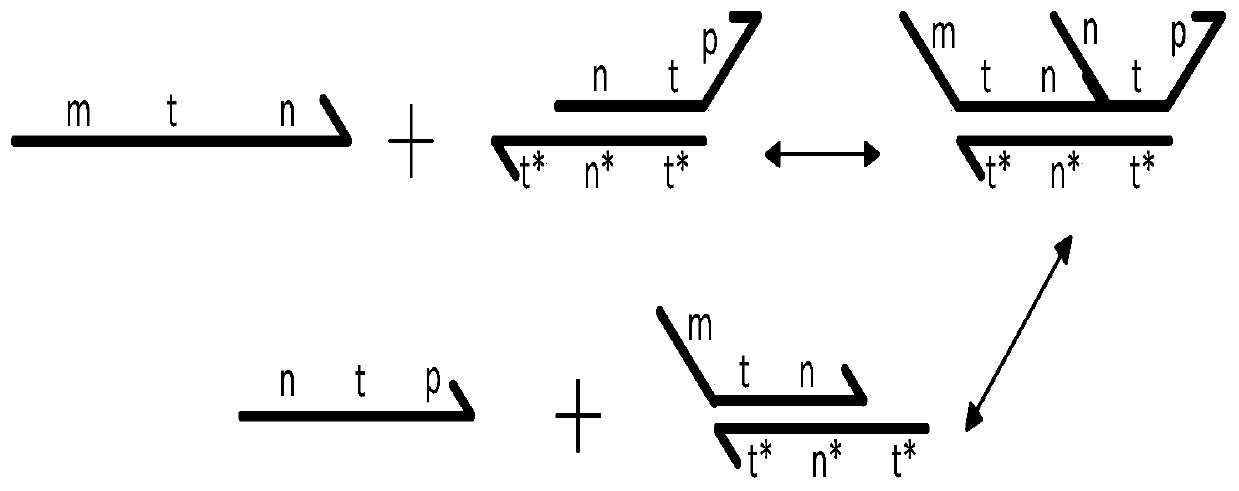
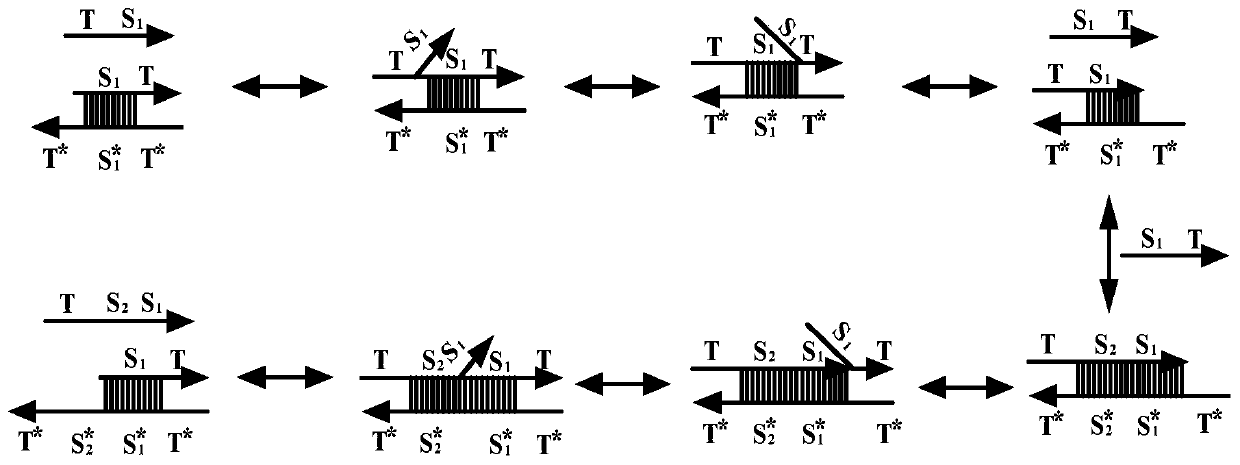
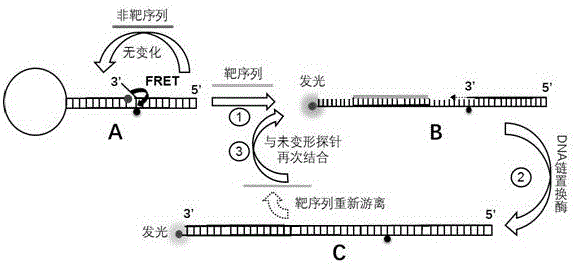
DNA strand displacement is a reaction where one of the strands in a double-stranded DNA is replaced with another nearly identical strand.
Color Image Encryption Method Based on DNA Strand Displacement Analog Circuit
InactiveUS20200287704A1A large amountShorten the timeKey distribution for secure communicationSecuring communication by chaotic signalsDna strand displacementA-DNA
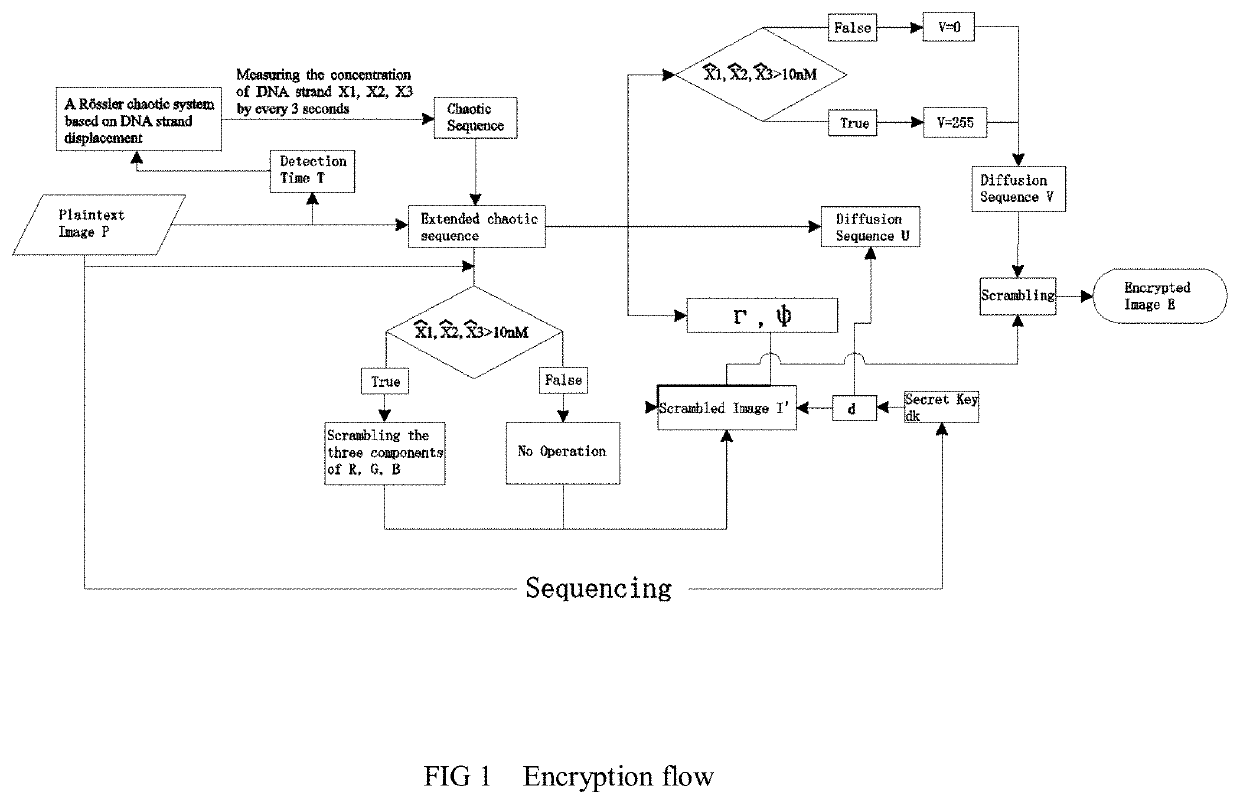
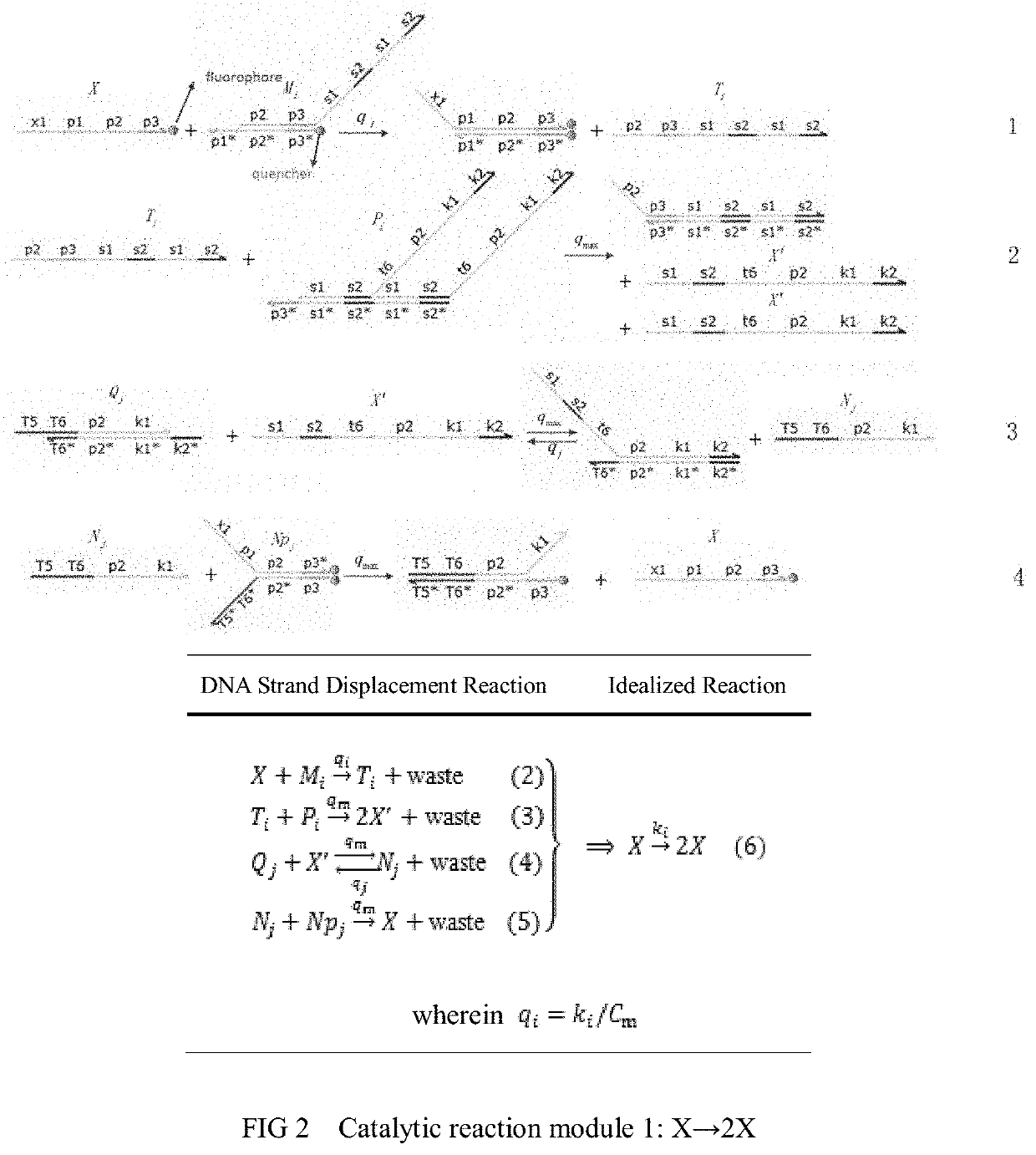
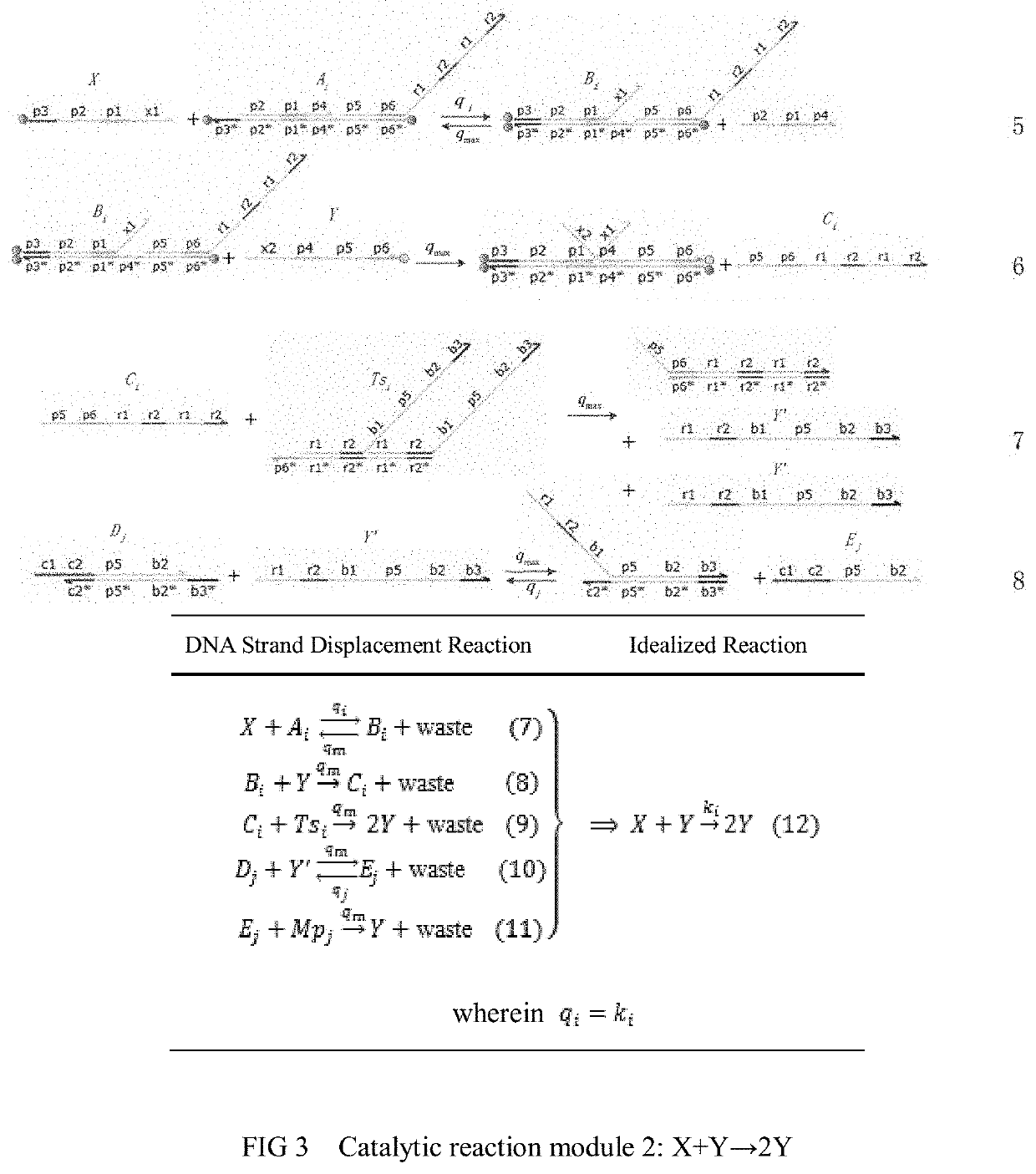
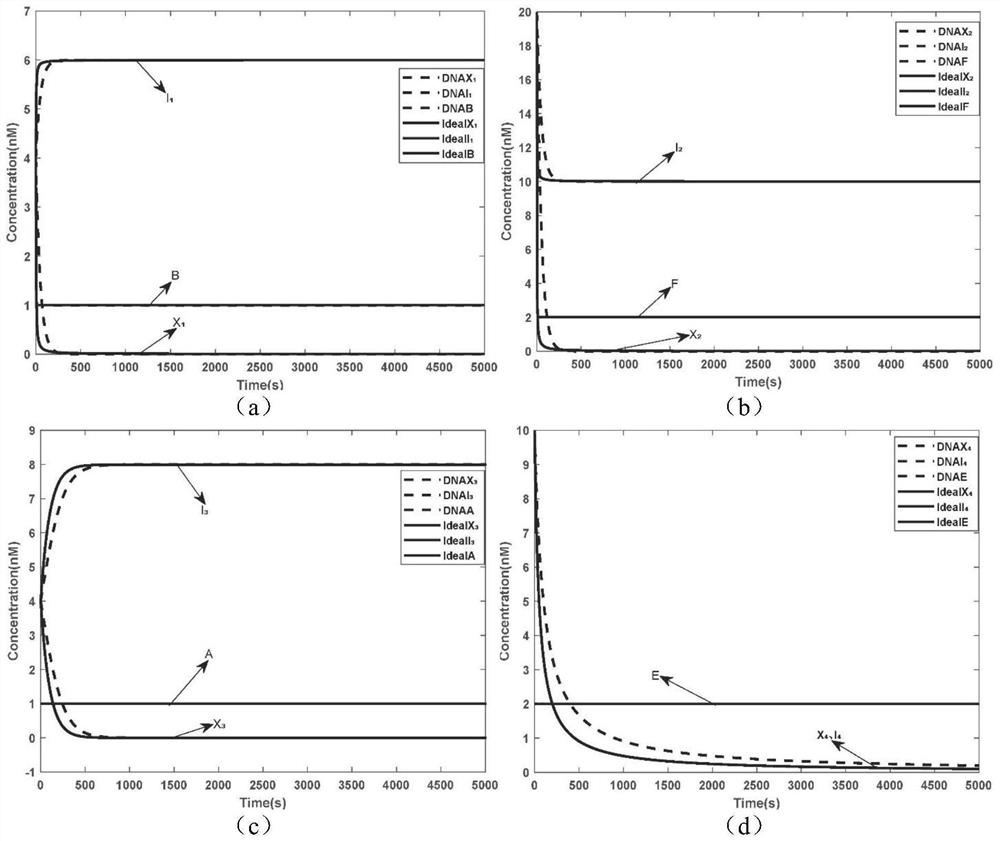
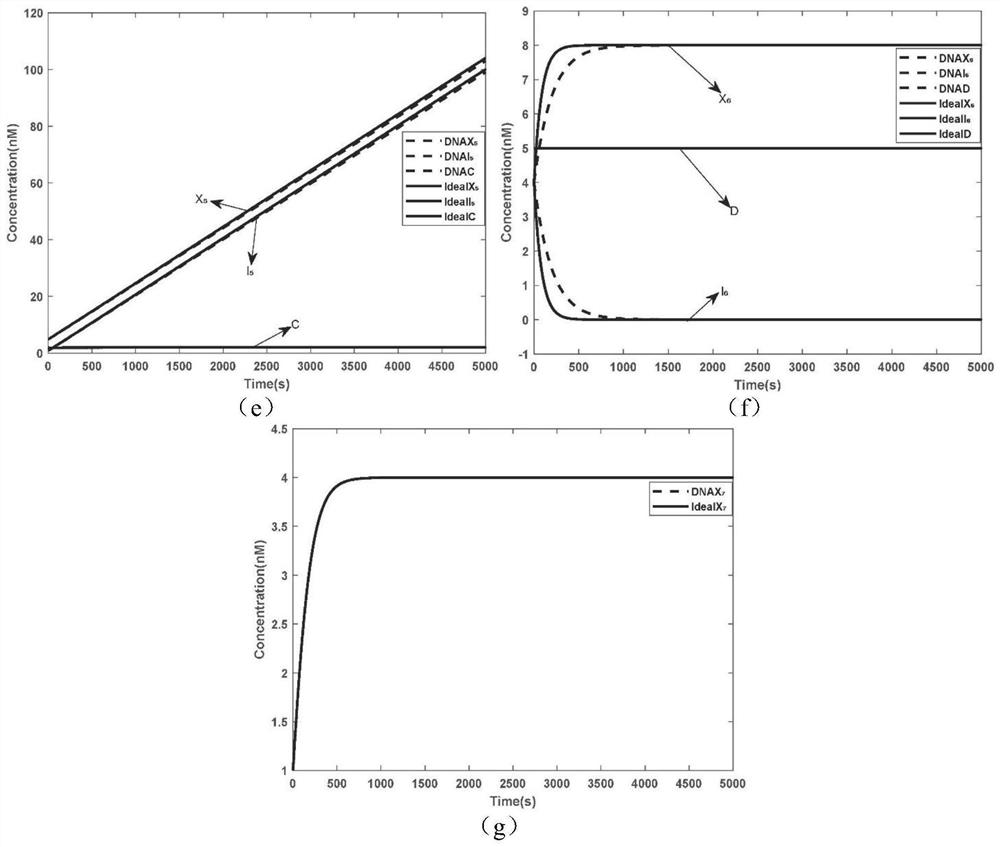
The invention relates to the field of strand displacement, and provides a color image encryption method based on a DNA strand displacement analog circuit. Firstly, a reaction module with a light-emitting group and a quenching group is designed through Visual DSD software, and by utilizing the equivalence of a DNA strand displacement reaction module and an ideal reaction module, an analog circuit formed by the DNA strand displacement reaction module can perform analog on the dynamics characteristics of an ideal reaction network formed by the ideal reaction module, wherein the Rössler chaotic system can be described by the idealized reaction network. Secondly, data generated by the DNA strand displacement analog circuit is converted into a chaotic sequence matched with a plaintext image in size after being extended, and finally, the color image encryption effect is achieved through scrambling and diffusion operations.
Owner:ZHANG QIANG +3
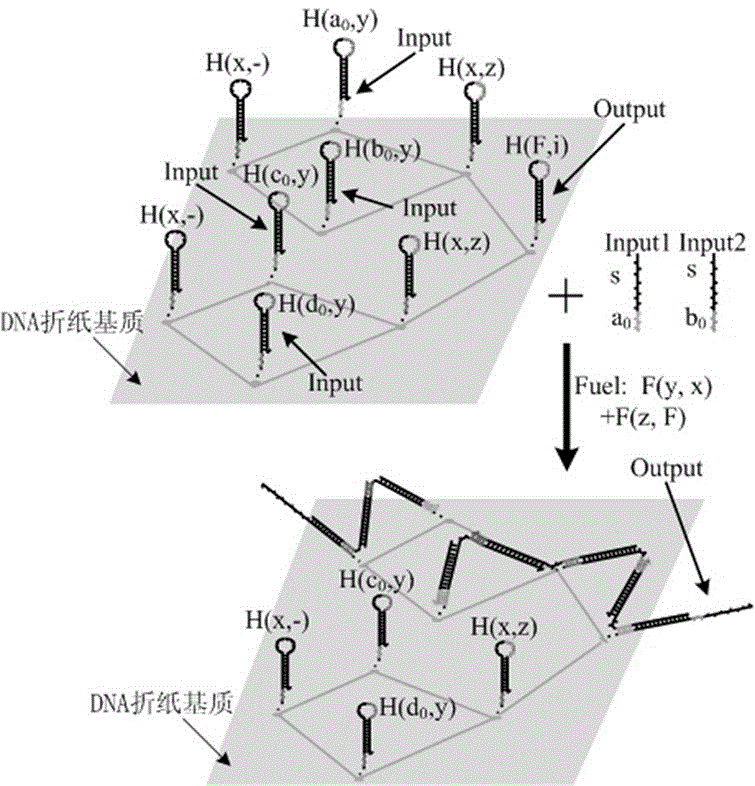
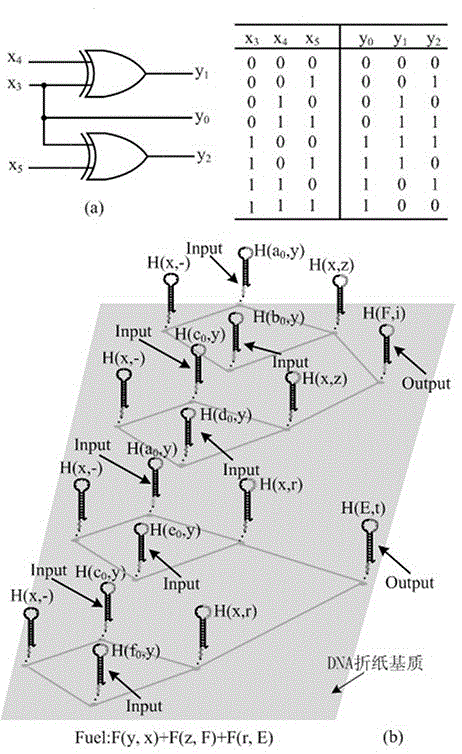
Local DNA hairpin strand displacement reaction-based XOR gate and complementing circuit
InactiveCN105930586AHigh precisionBuild fastCAD circuit designSpecial data processing applicationsDna strand displacementDisplacement reactions
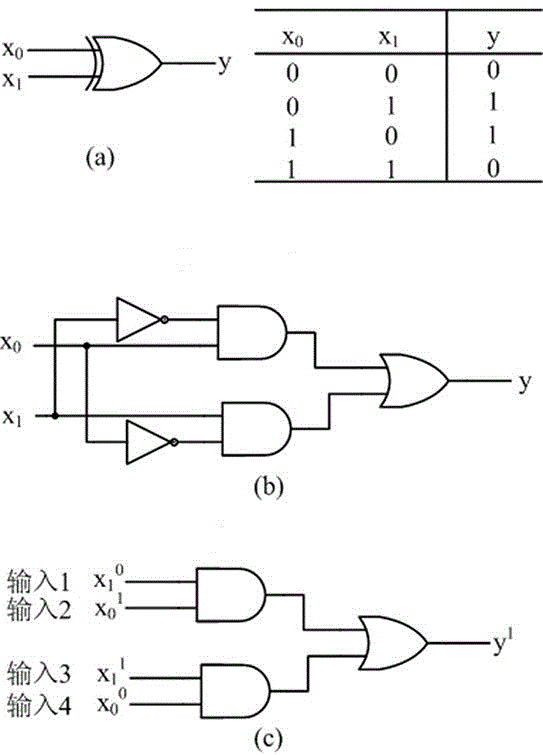
The invention belongs to the technical field of molecular circuit-based design and particularly relates to a DNA strand self-assembly principle-based and local DNA hairpin strand displacement reaction-based XOR gate and a complementing circuit constructed by utilizing the same. The XOR gate is constructed by adopting double-rail logic and comprises a single-stranded DNA input, a fuel hairpin and a DNA hairpin fixed to a fold paper substrate. The complementing circuit belongs to a three binary input complementing circuit and is composed of two parallel XOR gates. According to the provided XOR gate, the occurrence probability of wrong strand displacement reactions is relatively low and the DNA strand displacement reaction efficiency is improved. According to the provided complementing circuit, parallel computing can be realized without mutual interference; and a simulation result for Visual DSD of the complementing circuit shows that the complementing circuit is reasonable to construct, has good performance and has the advantages of high construction speed, high precision, high expandability and the like in construction of a large complex molecular system.
Owner:ZHENGZHOU UNIVERSITY OF LIGHT INDUSTRY
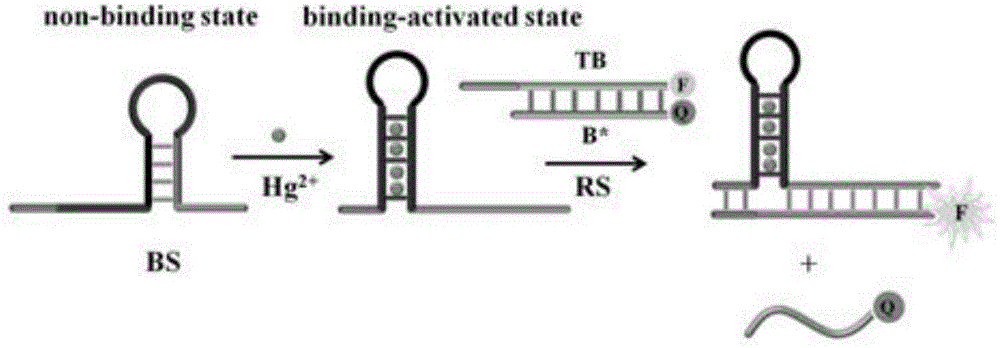
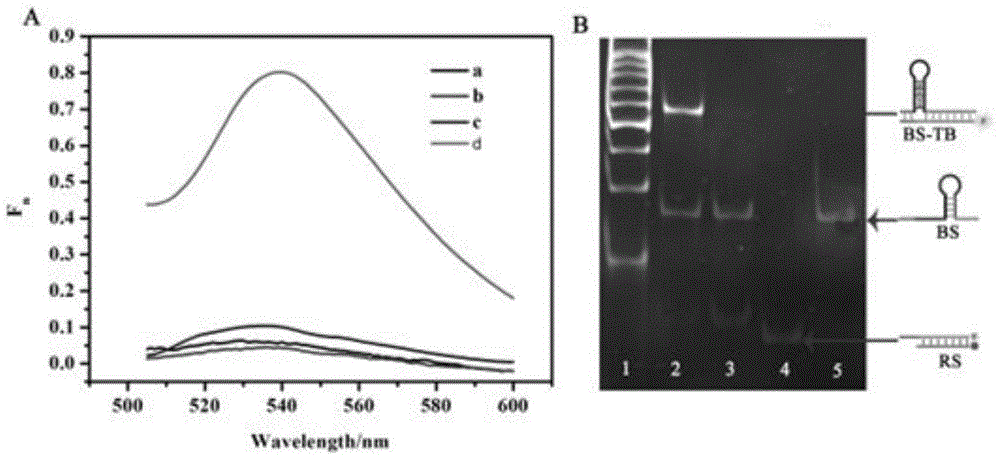
Suturing Toehold activation method used for controlling DNA strand displacement reaction and toolkit
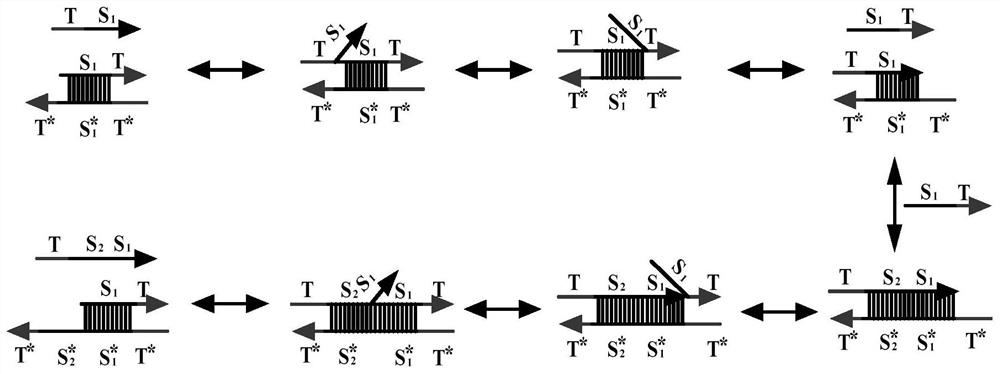
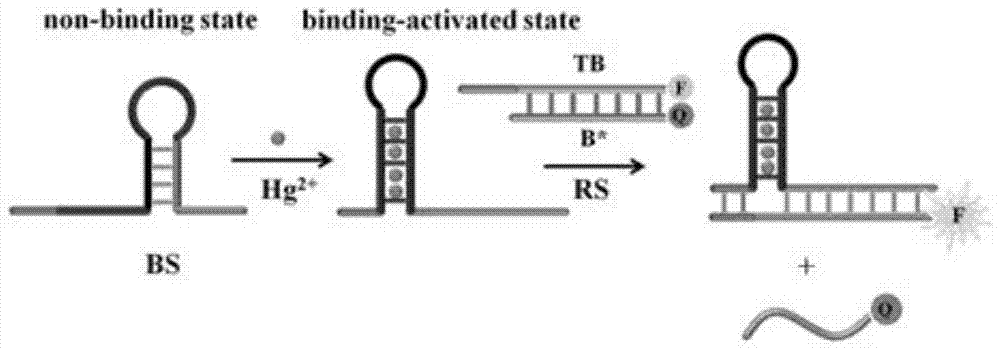
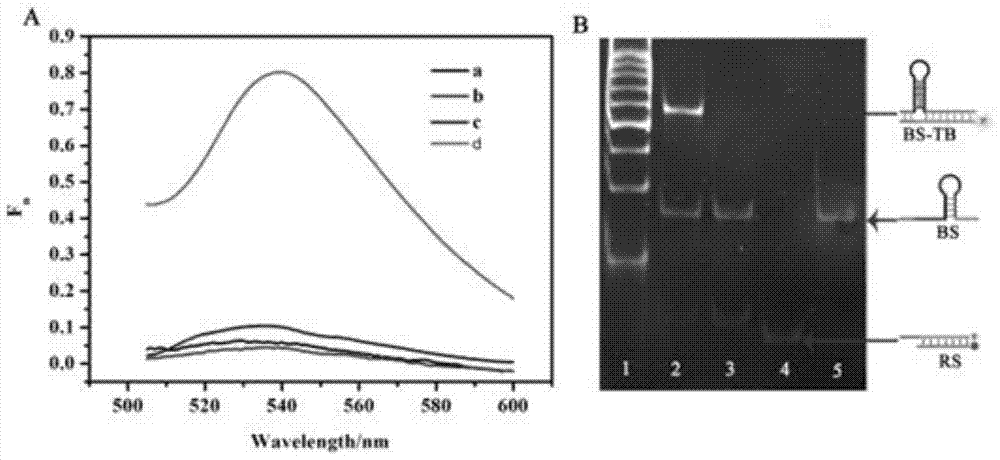
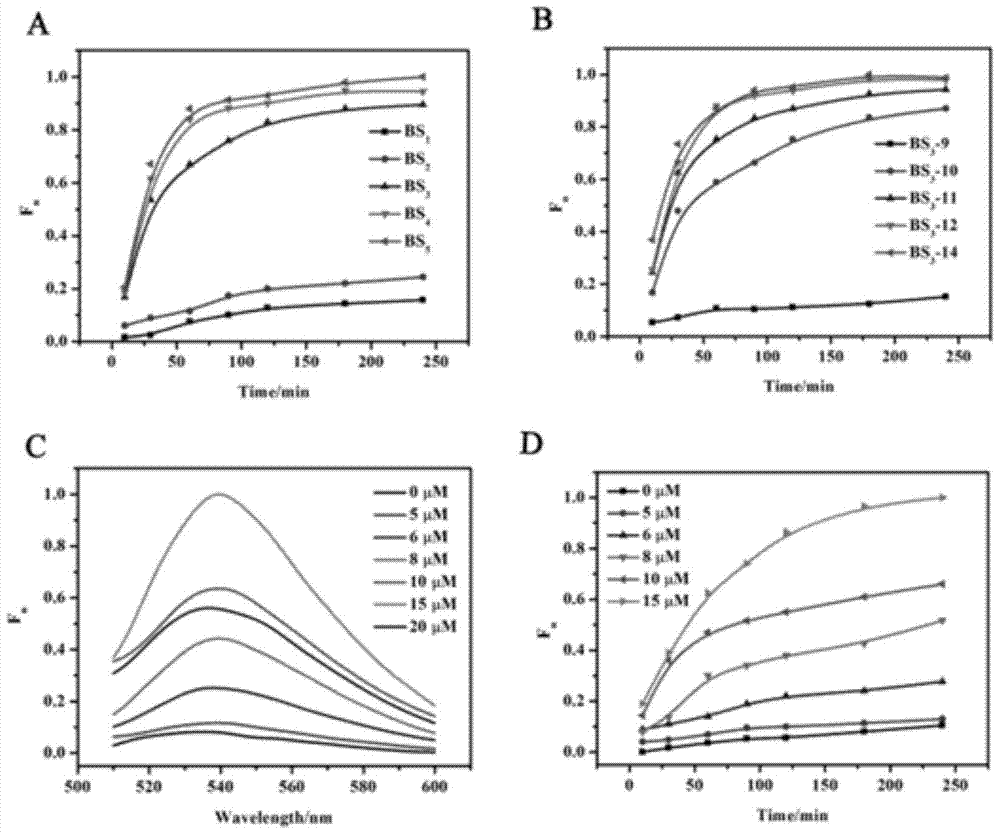
The invention provides a suturing Toehold activation method used for controlling a DNA strand displacement reaction and a toolkit. A novel Toehold activation method is developed based on hairpin reconstruction caused by mutual action of a DNA hairpin structure and different environmental irritants (Hg2+ or ATP). The invention develops a novel Toehold activation method. The extra level of the DNA strand displacement reaction can be accurately controlled by changing the concentration of the environmental irritants.
Owner:SHANDONG UNIV
Method for detecting bursaphelenchus xylophilus, detection primer and LAMP (loop-mediated isothermal amplification) detection kit of detection primer
ActiveCN103555840ASimple and fast operationStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationFluorescenceDna strand displacement
The invention discloses a method for detecting bursaphelenchus xylophilus, a detection primer and a LAMP (loop-mediated isothermal amplification) detection kit of the detection primer. The method for detecting the bursaphelenchus xylophilus comprises the steps of (1) extracting a DNA (deoxyribonucleic acid) of the bursaphelenchus xylophilus in a pine wood sample; (2) constructing a LAMP reaction system for loop-mediated isothermal amplification; (3) judging whether the bursaphelenchus xylophilus exists in the pine wood sample according to the change of fluorescence or turdity of an amplified product. The invention also provides a method which is easy to operate, low in cost and high in stability and is used for extracting the DNA of the bursaphelenchus xylophilus from pine wood. The invention further provides a LAMP primer group with high specificity and high sensitivity for detection of the bursaphelenchus xylophilus. The invention also provides the LAMP detection kit for the bursaphelenchus xylophilus. The LAMP detection kit comprises the LAMP primer group, a reaction buffering solution, a DNA chain replacement enzyme and a fluorescent dye / specific probe. The detection kit disclosed by the invention has the advantages of high specificity, high sensitivity, convenience and quickness in detection and the like.
Owner:杭州益森键生物科技有限公司
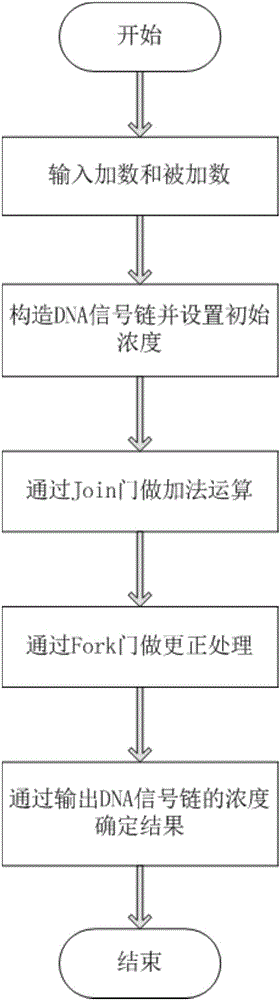
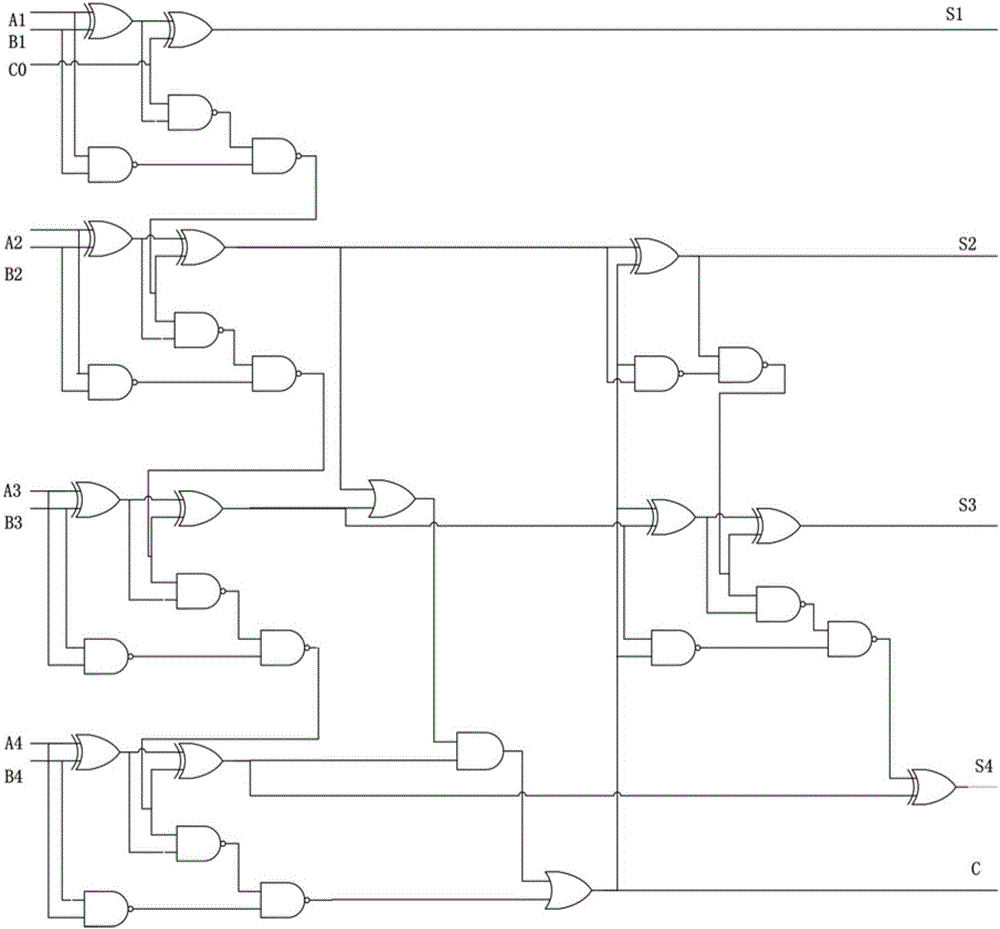
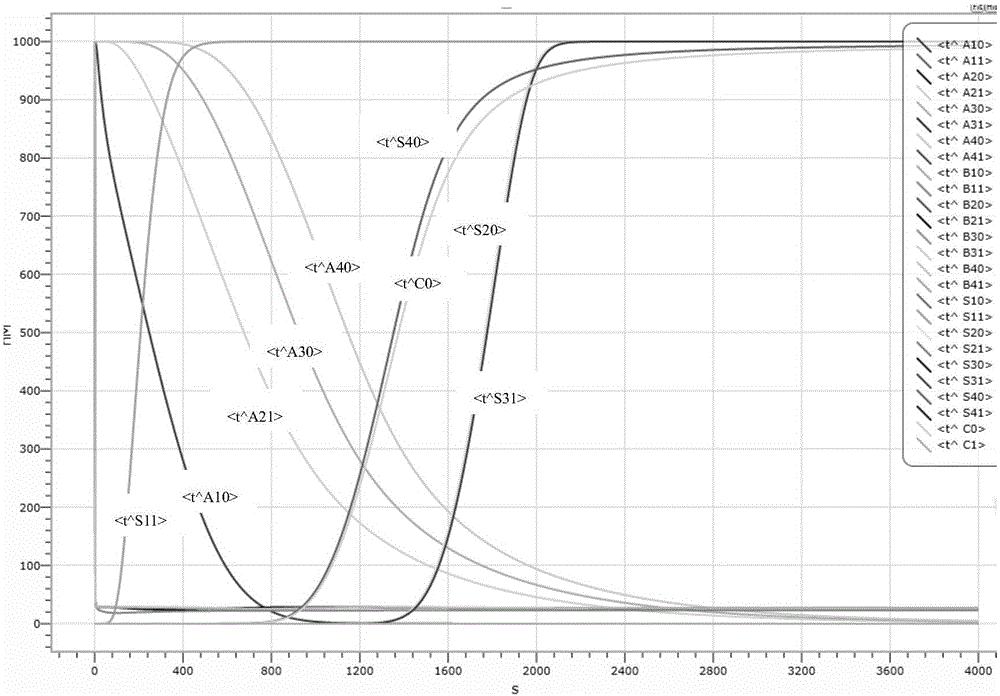
Design method of four-bit BCD code summator based on strand displacement
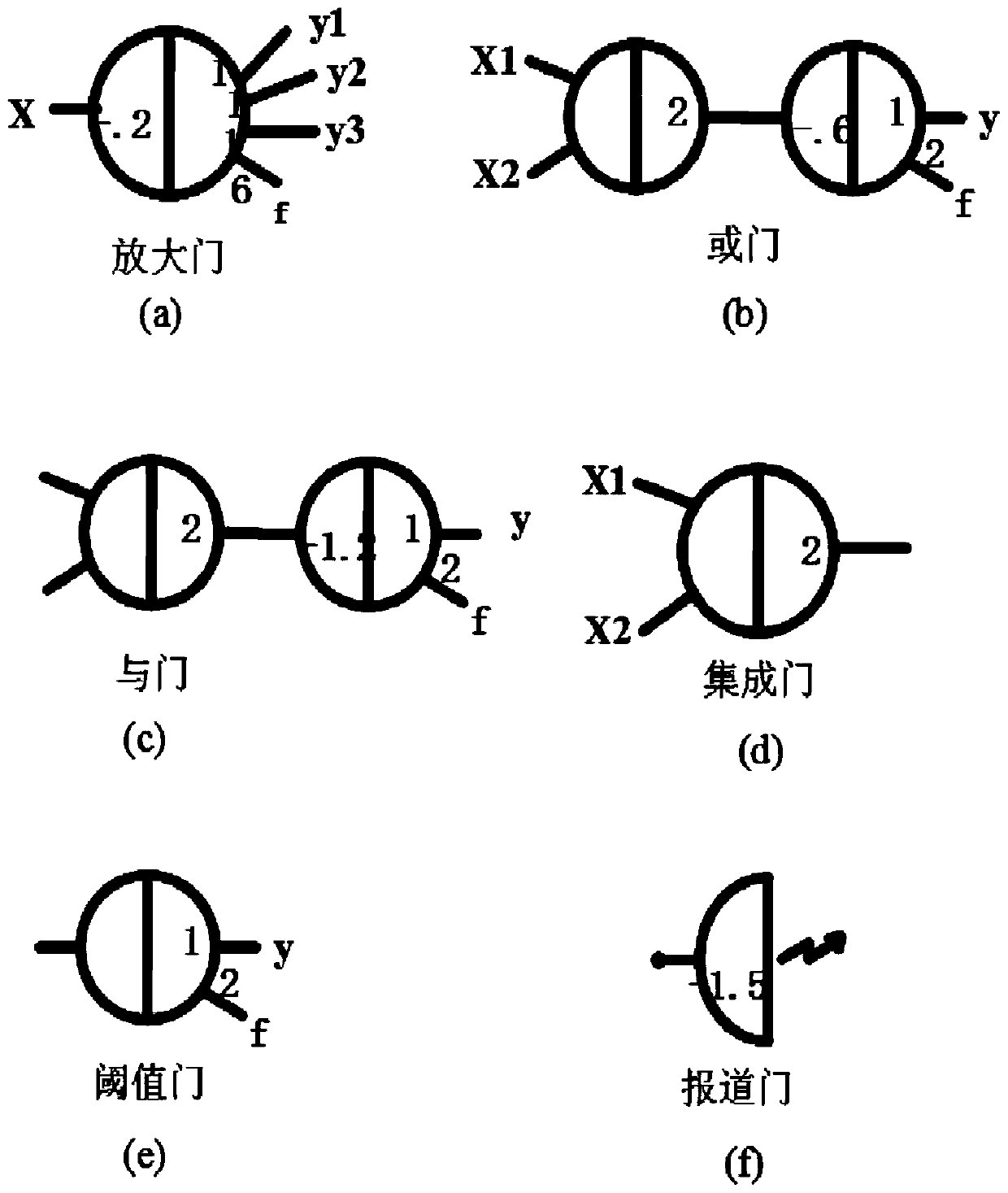
ActiveCN106650306AAccurate overflow judgmentThe displacement reaction is sufficientComputations using multiple devicesSpecial data processing applicationsNOR gateDna strand displacement
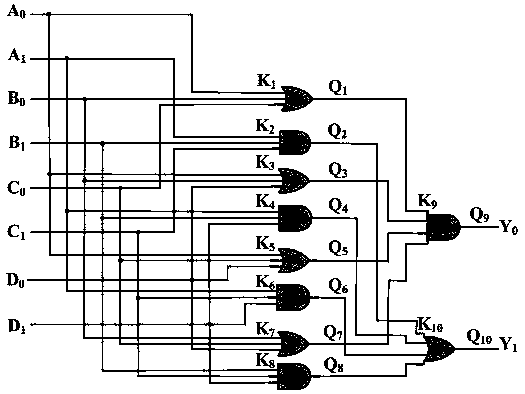
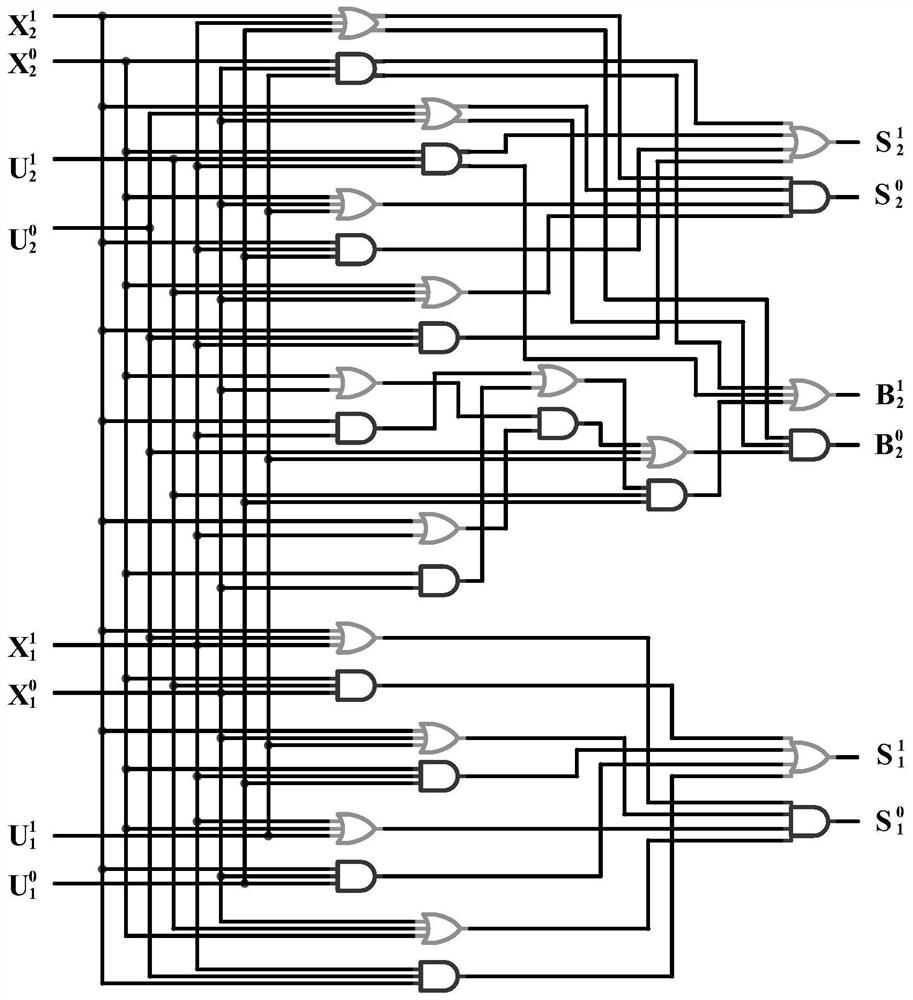
The invention relates to the field of strand displacement, and discloses a design method of a four-bit BCD code summator based on DNA strand displacement. In the design method, a thought of double rail logic is introduced, a monorail logic circuit comprising 17 alternative denial gates, 12 nor gates, 2 or gates and 1 and gate is simulated through a DNA strand and is converted into a double rail logic to be participate in a reaction. The method ensures that reaction is more stable and efficient, and logic of consequence obtained is clearer and easy to understand. From the view of experimental data and experimental results, the method not only can correctly process carry bit and overflow during operation, but also is high in efficiency and stable in result, and explains the design effectiveness of the summator.
Owner:DALIAN UNIVERSITY
Fire alarm dual-rail logic circuit based on strand displacement, and implementation method thereof
ActiveCN108233919AImprove reliabilityHigh sensitivityLogic circuits characterised by logic functionDesign optimisation/simulationSmoke detectorsSystems design
The invention provides a fire alarm dual-rail logic circuit based on strand displacement, and an implementation method thereof. Based on a reaction mechanism of DNA strand displacement, a four-input fire alarm circuit system is constructed, and a digital logic circuit for fire alarm circuit operation capable of inputs of a smoke detector, a temperature detector, a light detector and a manual alarmdevice is erected; the digital logic circuit is transformed into a dual-rail logic circuit through the dual-rail logic idea; the dual-rail logic circuit is then transformed into a seesaw biochemicallogic circuit; and an output result is validated through Visual DSD simulation software, and whether a fire occurs or not is analyzed and judged. The circuit and the method provided by the invention have the advantages that the circuit is effectively designed and is highly accurate and sensitive; and a basic theoretical foundation is provided for large-scale intelligent alarm device system designing operation, so that a bio-computer logic circuit is more reliable, and the development of bio-computers is promoted.
Owner:ZHENGZHOU UNIVERSITY OF LIGHT INDUSTRY
Method for implementing single-molecule chemical reaction network by DNA (deoxyribonucleic acid) strand displacement reaction
ActiveCN108009395AReduce complexityChemical processes analysis/designComputational theoretical chemistryChemical reactionDisplacement reactions
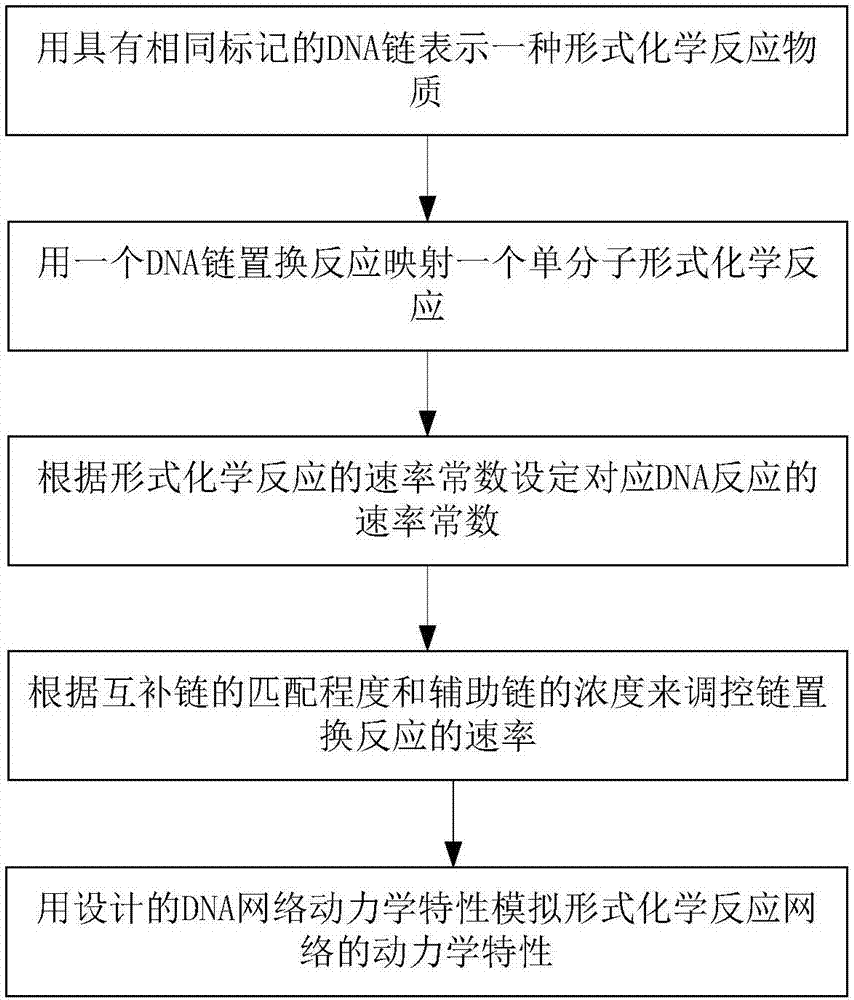
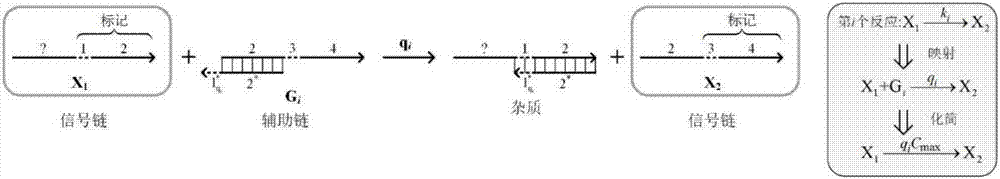
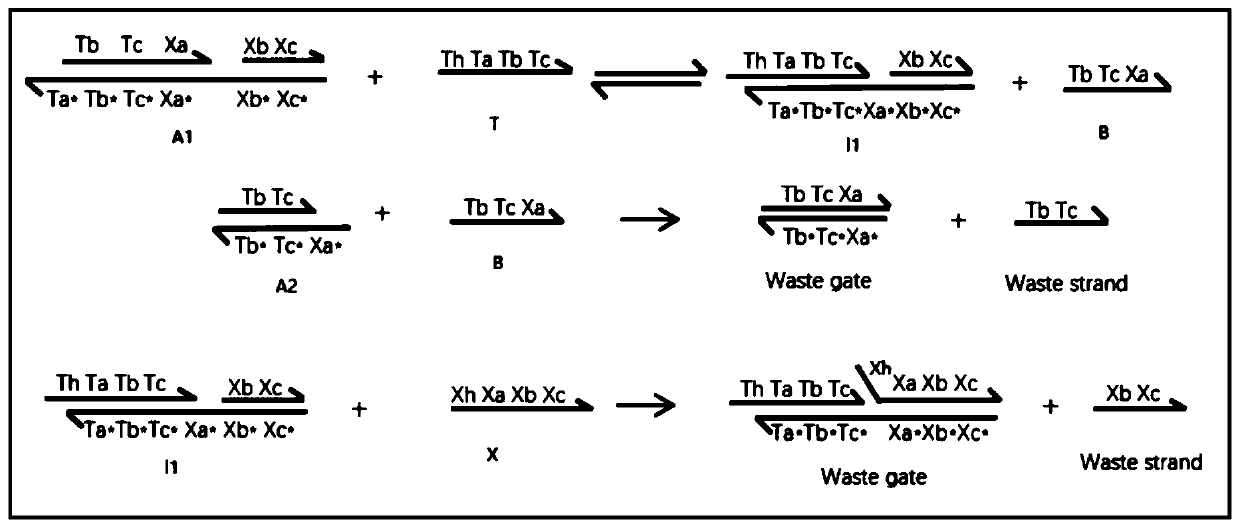
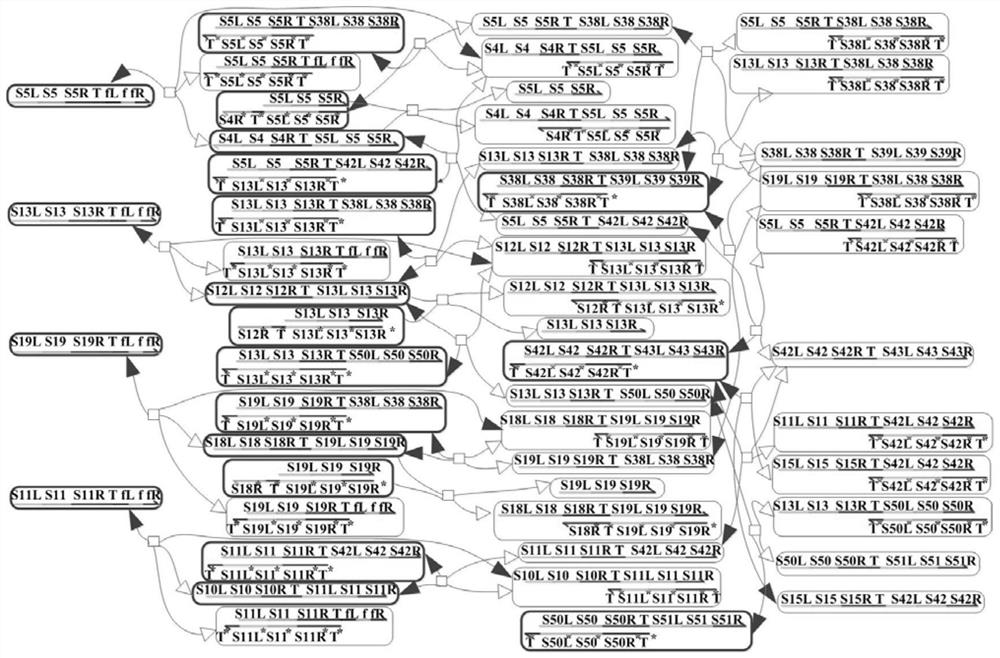
The invention discloses a method for implementing a single-molecule chemical reaction network by DNA (deoxyribonucleic acid) strand displacement reaction. The method includes the steps: S1 expressingone chemical reaction substance by the aid of DNA strands with same marks; S2 mapping one irreversible single-molecule chemical reaction by the aid of one DNA strand displacement reaction, and mappingone reversible single-molecule chemical reaction by the aid of two DNA strand displacement reactions; S3 setting a rate constant of the corresponding DNA reaction according to the a rate constant ofthe chemical reaction; S4 adjusting the rate of the strand displacement reaction according to the matched degree of a short strand in mark strands and a complementary strand of the short strand; S5 simulating dynamic characteristics of the chemical reaction network by the aid of dynamic characteristics of a DNA network formed by all DNA strand displacement reactions. Each mark comprises a short strand and a long strand, a reaction network formed by a plurality of chemical reactions is a chemical reaction network, and the chemical reaction of the corresponding DNA strand reaction multiplies initial concentration of an auxiliary strand to be equal to the rate constant of the chemical reaction.
Owner:SOUTHEAST UNIV
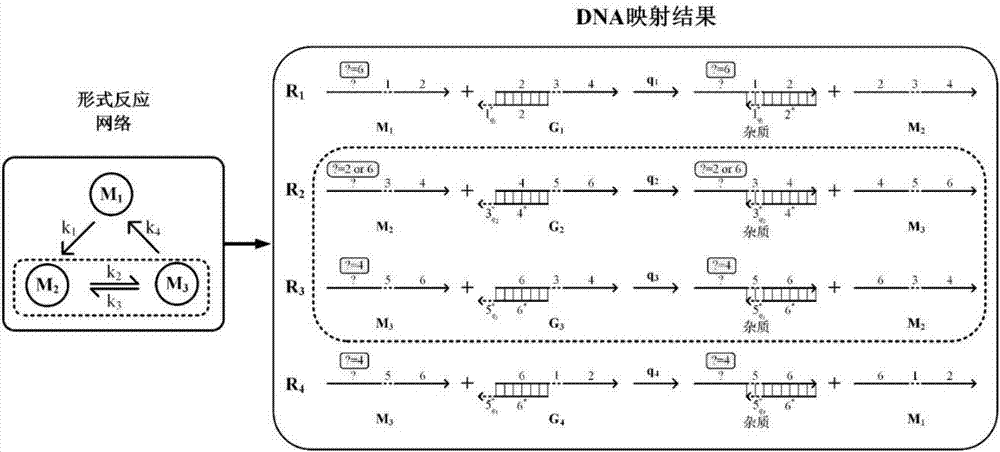
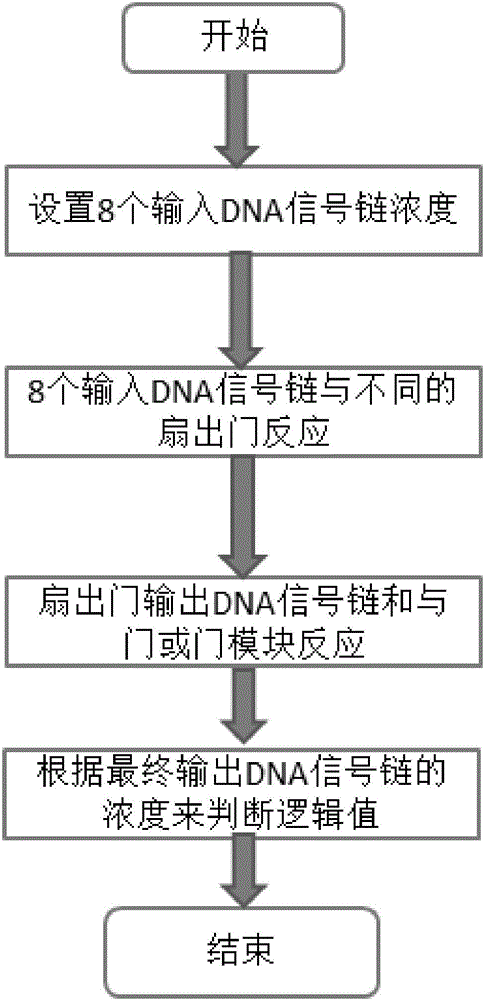
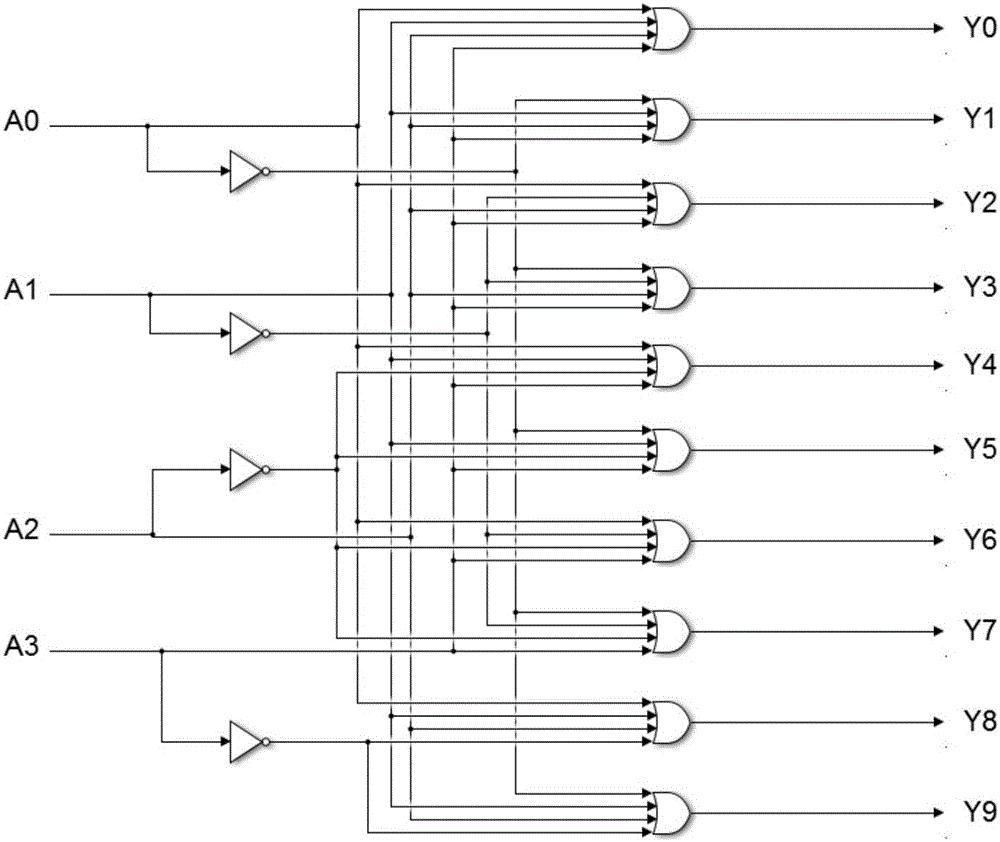
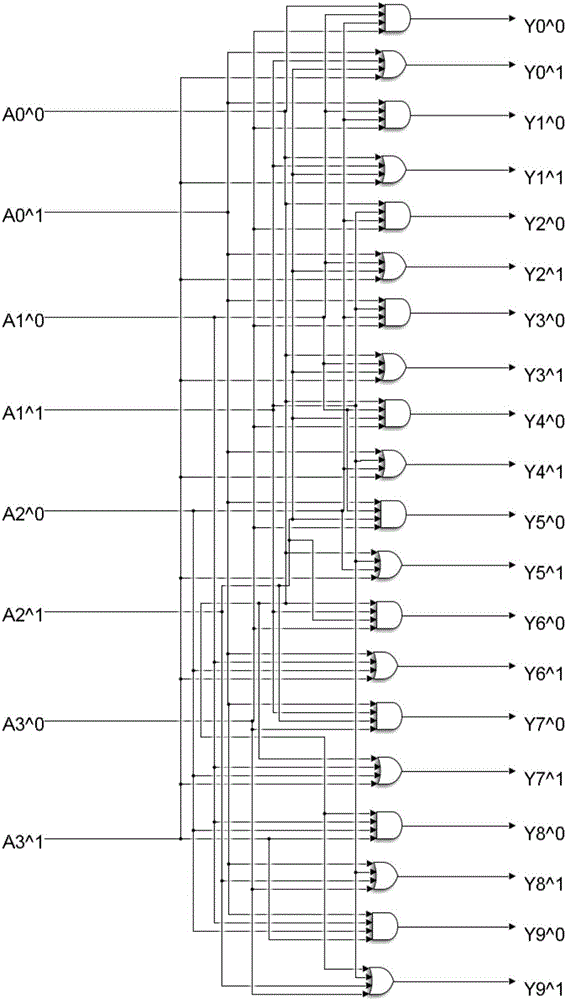
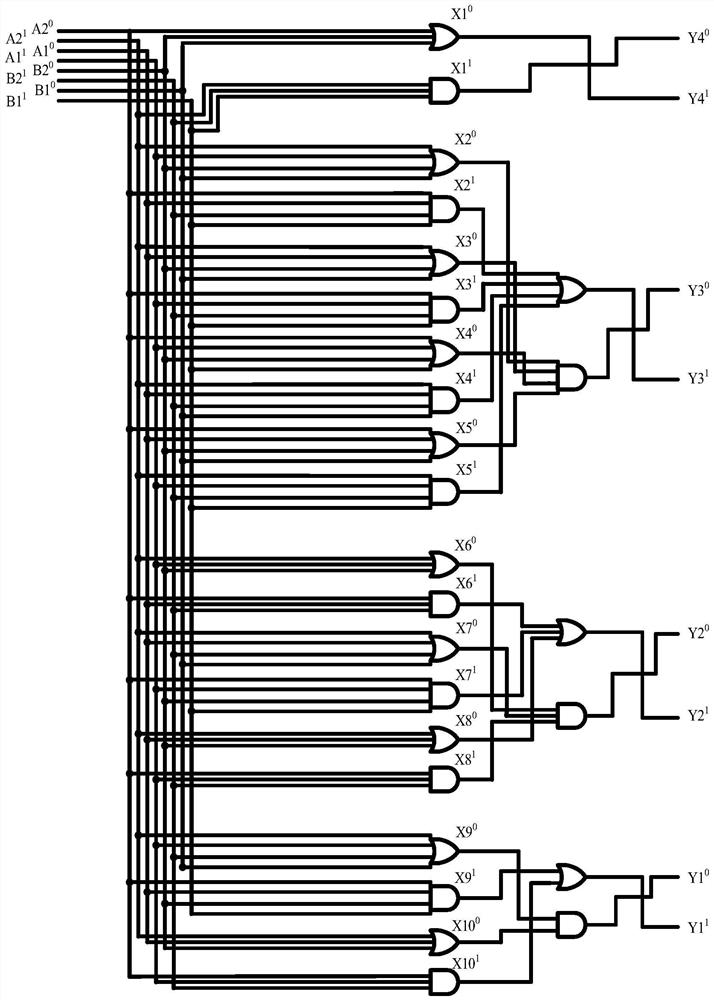
4-10 decoder design method based on strand displacement
The invention provides a 4-10 decoder design method based on strand displacement, and relates to the field of DNA strand displacement. According to the method, firstly concentration of DNA signal chains is set according to the input logic value of a DNA 4-10 decoder, and each DNA signal is divided into 10 different DNA signal chains having the same logic value through a fan-out gate module; then hybridization reaction with an AND gate module and an OR gate module is performed based on the Watson-Crick (C-G, T-A) complementary matching principle according to difference of domains attached on the outputted 80 DNS signal chains; and finally 20 DNS signal chains are outputted, and the 20 DNS signal chains represent different logic values according to the size of concentration so that the function of the 4-10 decoder can be realized. The beneficial effects of the 4-10 decoder design method based on strand displacement are that the AND gate module, the OR gate module and the fan-out gate module are combined together based on double tracks firstly so that the DNA 4-10 decoder is realized; and the output result is enabled to be more stable by regulating the concentration coefficient of the threshold gate in the fan-out gate module, the AND gate module and the OR gate module.
Owner:DALIAN UNIV
SERS-fluorescent dual mode probe based on DNA chain replacement, and preparation method and application of SERS-fluorescent dual mode probe
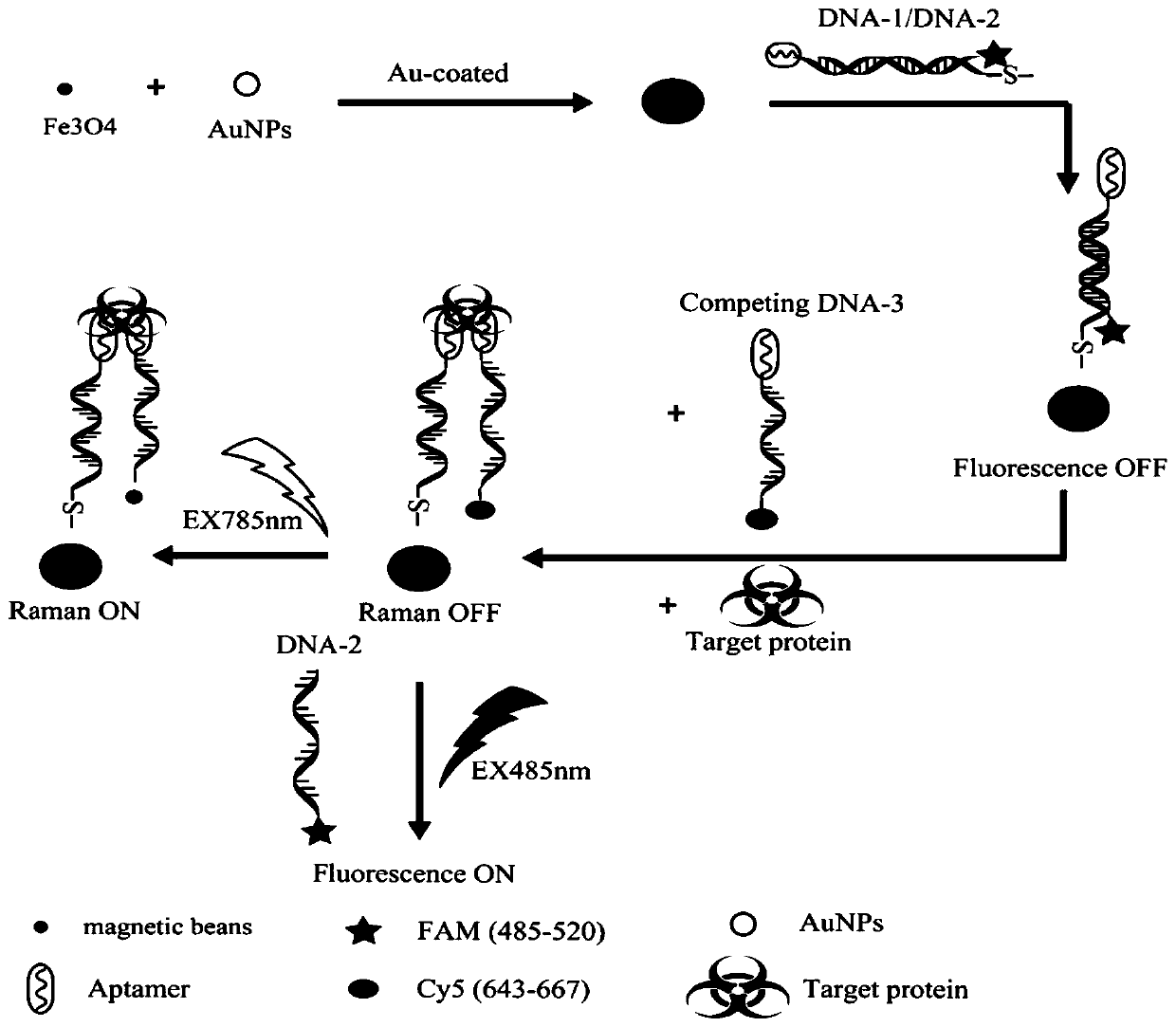
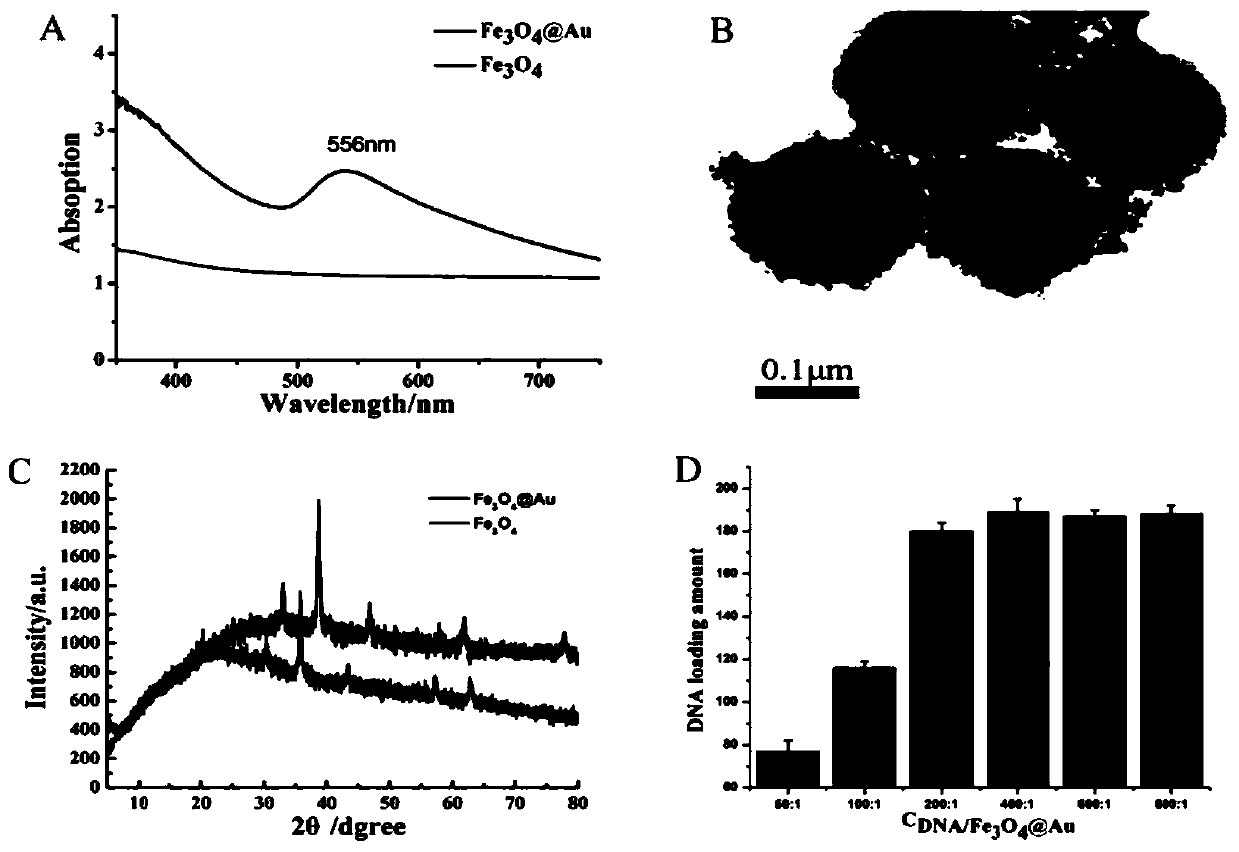
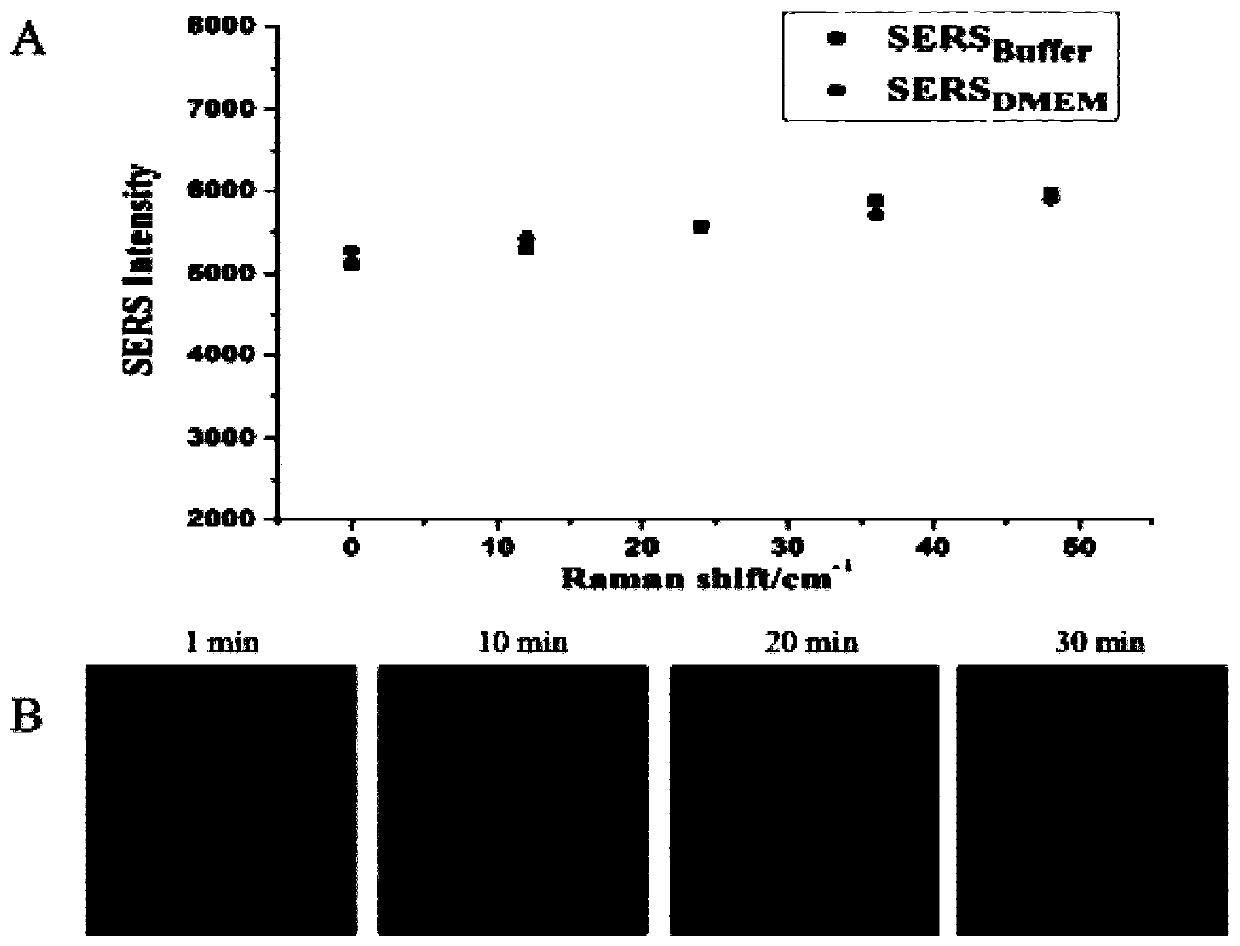
ActiveCN110387402ARealize detectionStable in natureMicrobiological testing/measurementAgainst vector-borne diseasesAptamerFluorescence
The invention discloses an SERS-fluorescent dual mode probe based on a DNA chain replacement principle. The SERS-fluorescent dual mode probe is prepared from a DNA1-2 double strand chain modified by aferroferric oxide magnetic composite material with a certain particle size and another DNA3 single strand chain; and the DNA1 and the DNA3 contain the same aptamer, the tail end of the DNA2 is modified with a fluorogen FAM6, and the tail end of the DNA3 is modified with a fluorogen Cy5. The invention further discloses a preparation method and application of the SERS-fluorescent dual mode probe based on the DNA chain replacement principle. According to the SERS-fluorescent dual mode probe based on the DNA chain replacement principle, two types of signals of raman and fluorescence can be used for simultaneously detecting biomarkers in superficial living cells, detecting is fast, convenient and efficient, the specificity is high, the sensitivity is high, and the accuracy is high.
Owner:SUN YAT SEN UNIV
Method for detection of a nucleic acid target sequence
Disclosed is a method of facilitating detection of a nucleic acid target sequence. The method may include obtaining a toehold-mediated DNA strand displacement apparatus comprising a portion complementary to the nucleic acid target sequence. The method may further include obtaining an RNA toehold switch comprising an RNA sequence comprising an inverted loop with a sequestered ribosome binding site and a sequestered translation start codon, a transducer sequence on the 3′ side of the inverted loop, coding for a reporter protein, and a toehold portion on the 5′ side of the inverted loop. Further, the toehold portion of the RNA sequence may be complementary to a portion of the Toehold-mediated DNA strand displacement apparatus. The method may further include combining the Toehold-mediated DNA strand displacement apparatus and the RNA toehold switch in an assay, such that the two are never in direct physical contact with each other. Accordingly, a sample containing the nucleic acid target sequence on the substrate may displace a nucleic acid strand from the toehold-mediated DNA strand displacement apparatus and bind a portion of it to the RNA toehold switch resulting in expression of the reporter protein.
Owner:TOTEMID INC
Implementation method of three-cascade molecular combinational circuit based on DNA strand displacement
ActiveCN110533155AProof of validityImprove reliabilityLogic circuits characterised by logic functionBiomolecular computersDna strand displacementDisplacement reactions
Owner:ZHENGZHOU UNIVERSITY OF LIGHT INDUSTRY
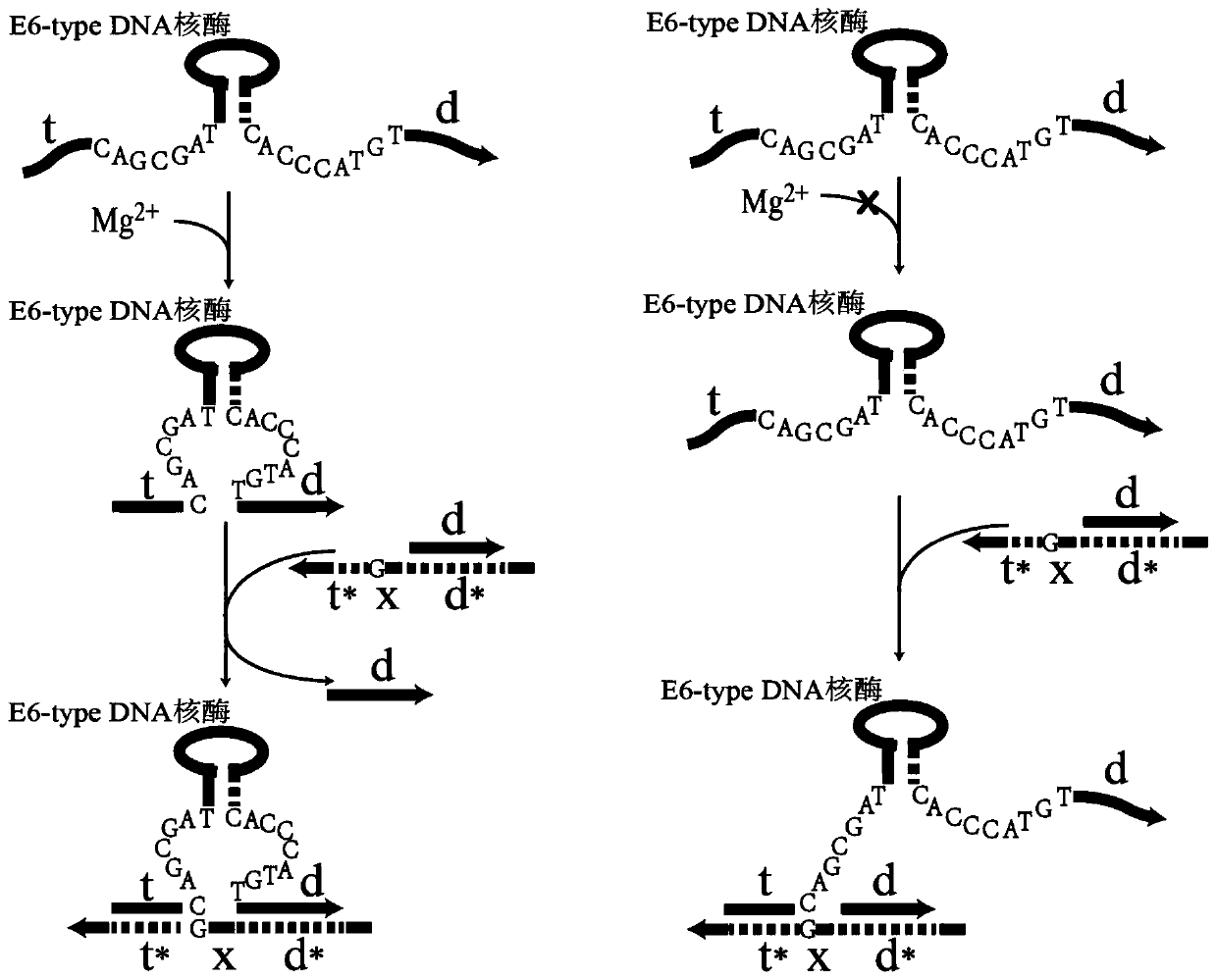
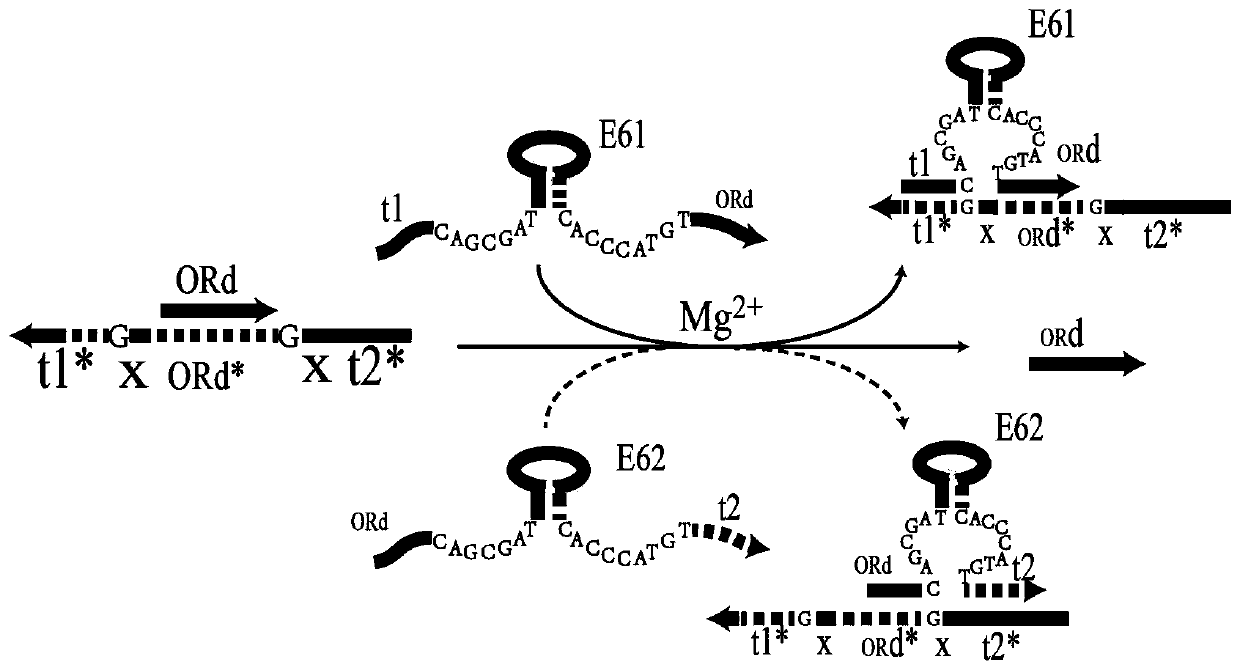
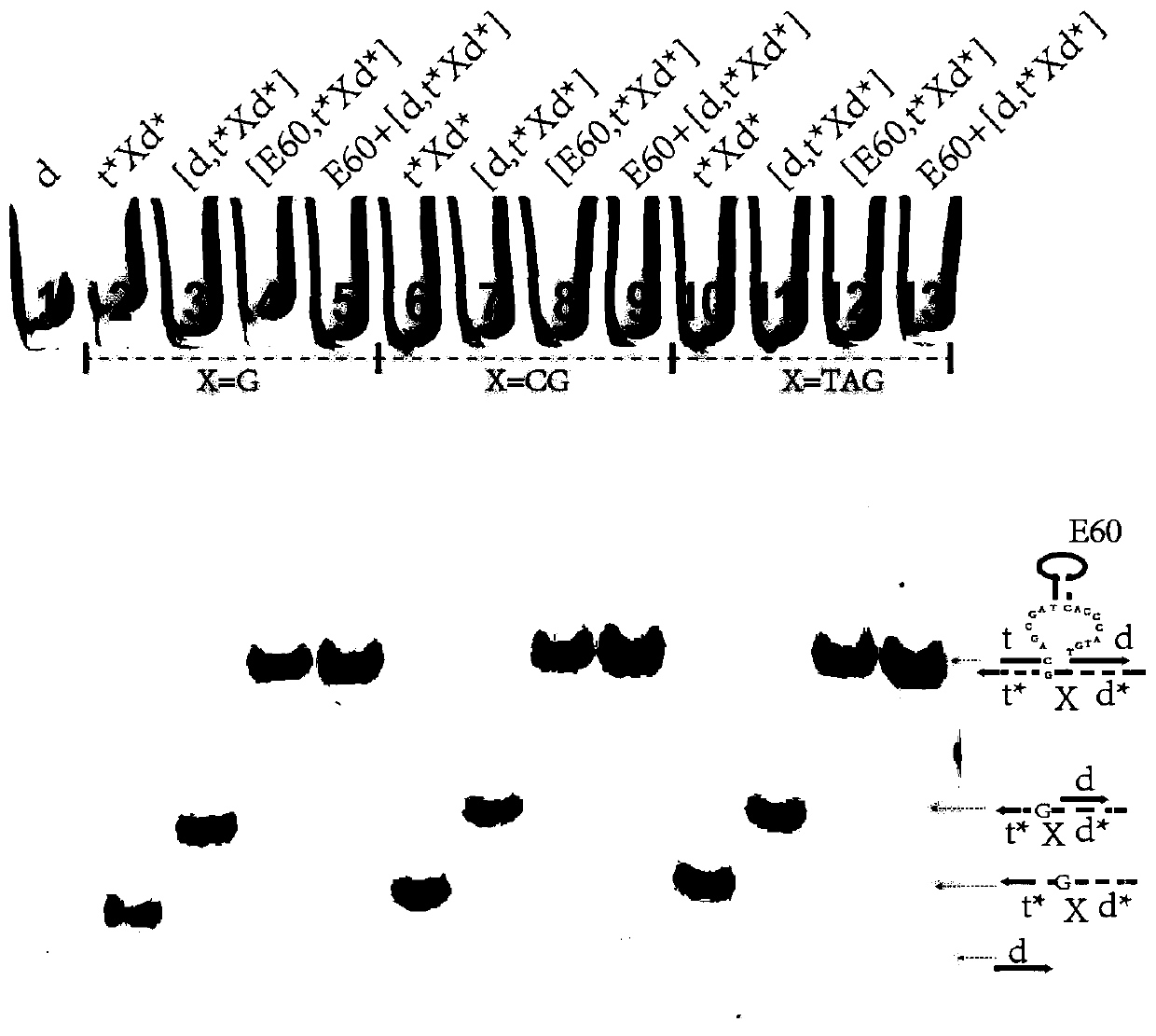
Remote DNA strand displacement method of magnesium ion induced E6-type DNA ribozyme allosterism
ActiveCN109797181AAchieve replacementIncrease flexibilityFermentationSequence designDna strand displacement
The invention relates to the field of molecular computing, and particularly relates to DNA strand displacement technology, in particular to a remote DNA strand displacement method of magnesium ion induced E6-type DNA ribozyme allosterism. In DNA strand displacement, a standpoint and a branch migration domain are separated and are not necessarily adjacent to each other in DNA sequence design; metalions are utilized to induce DNA strand displacement. A new DNA strand displacement control method is provided, remote DNA strand displacement is realized, and flexibility of DNA strand displacement is improved.
Owner:SHENYANG AEROSPACE UNIVERSITY
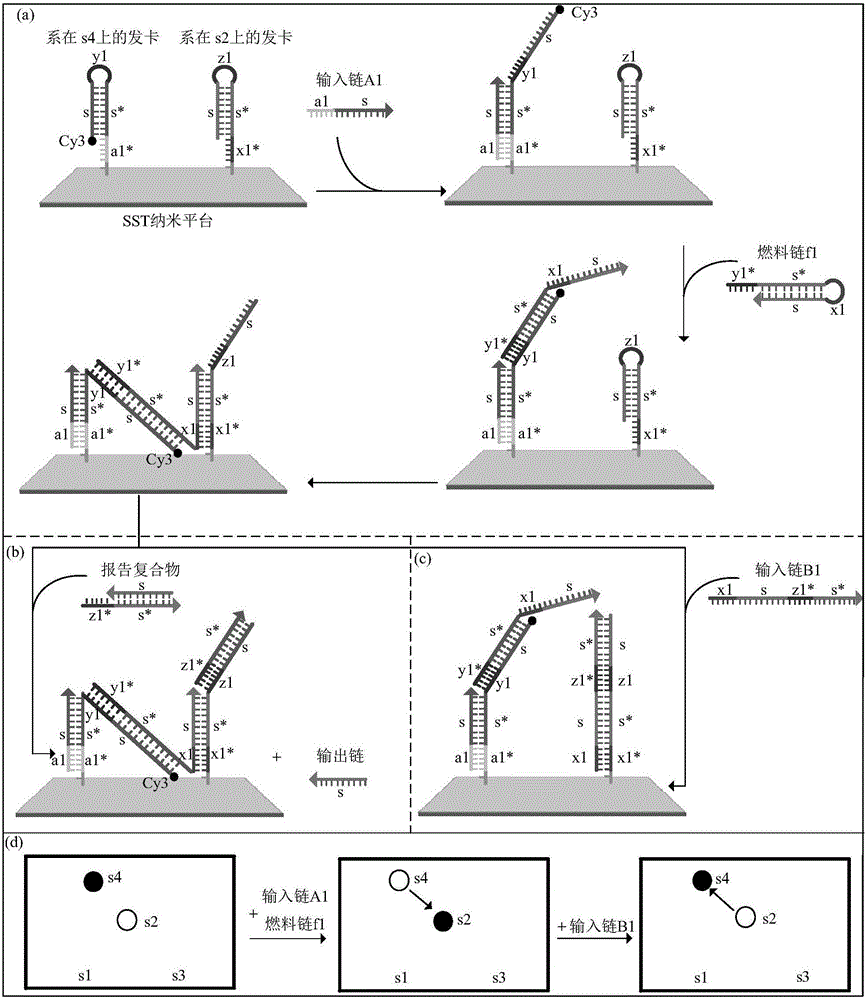
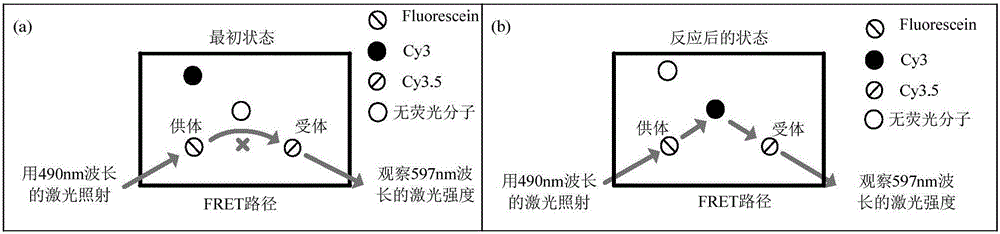
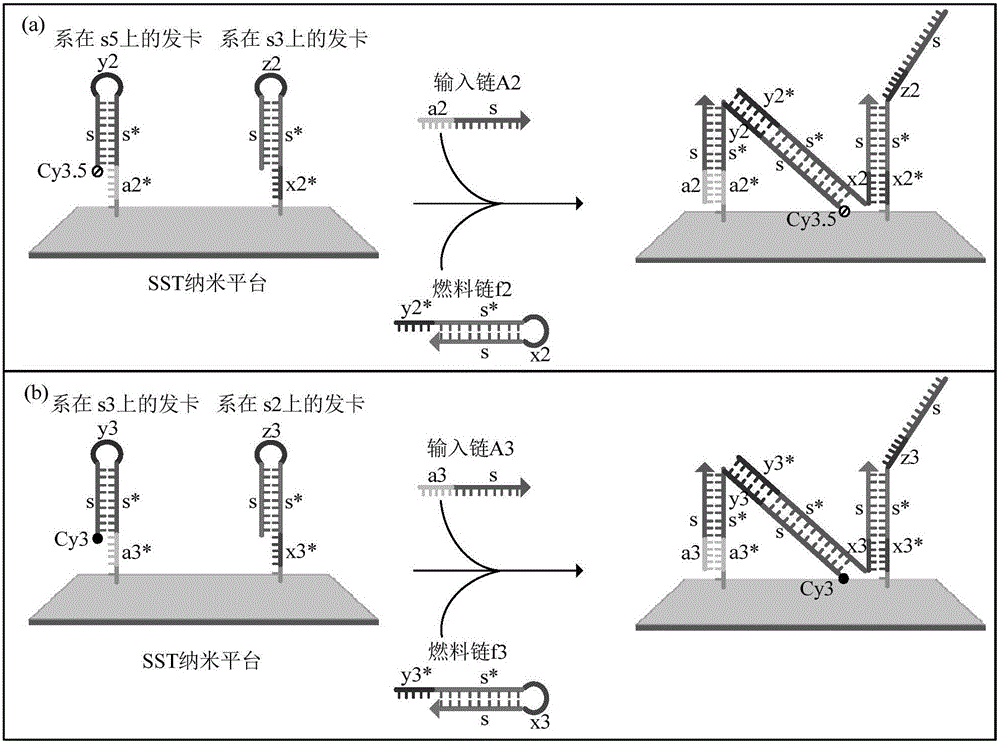
Phototonus logical circuit design method based on SST nanometer platform
ActiveCN105969869ARealize the logic circuitMicrobiological testing/measurementDNA nanotechnologyLogic circuit design
The invention belongs to the technical field of DNA nanometer, and relates to a phototonus logical circuit design method based on an SST nanometer platform. Fluorescent molecules are arranged on the SST nanometer platform through DNA self-assembly, the position of the fluorescent molecules is arrayed in the DNA chain displacement process, and a fluorescent molecule energy transfer path is formed among the reset fluorescent molecules. In serial design of the SST nanometer structure, a single chain taking part in follow-up DNA strand displacement reaction needs to extend a hairpin structure region, the extended sequence and specific system input chains take part in the strand displacement reaction, fluorescent molecule transfer systems are successfully built, and therefore the fluorescent molecules can be transferred at normal temperature. Configuration among the fluorescent molecules is controlled by uniting multiple fluorescent molecule transfer systems, and a phototonus molecular logical circuit is obtained by means of fluorescence resonance energy transfer technology.
Owner:郑州轻大产业技术研究院有限公司
Four-input factorial additive operation molecular circuit design method based on DNA strand displacement
ActiveCN110544511AImprove reliabilityComputation using non-contact making devicesSpecial data processing applicationsComputer architectureRandom combination
The invention provides a four-input factorial addition operation molecular circuit design method based on DNA strand displacement, and the method comprises the following steps: listing factorial multiplications corresponding to two binary numbers, listing a truth table of the sum of the factorial multiplications of ten kinds of two binary numbers according to a random combination, and constructinga factorial addition operation circuit; converting a four-input factorial additive operation circuit into a double-track logic circuit only comprising a logic AND gate and a logic OR gate by adoptinga double-track idea; designing a DNA molecular logic gate through DNA molecules, constructing a four-input fractional addition operation molecular circuit, verifying the correctness of an output result through Visual DSD simulation software, analyzing complex dynamic behaviors of the four-input fractional addition operation molecular circuit, and verifying the dynamic behaviors of the four-inputfractional addition operation molecular circuit. According to the invention, a basic theoretical basis is provided for constructing a more complex logic operation circuit in the future, and the development of the biological computer is promoted, so that the reliability of the logic circuit of the biological computer is improved.
Owner:ZHENGZHOU UNIVERSITY OF LIGHT INDUSTRY
Implementation method of two-bit Gray code subtractor molecular circuit based on DNA strand displacement
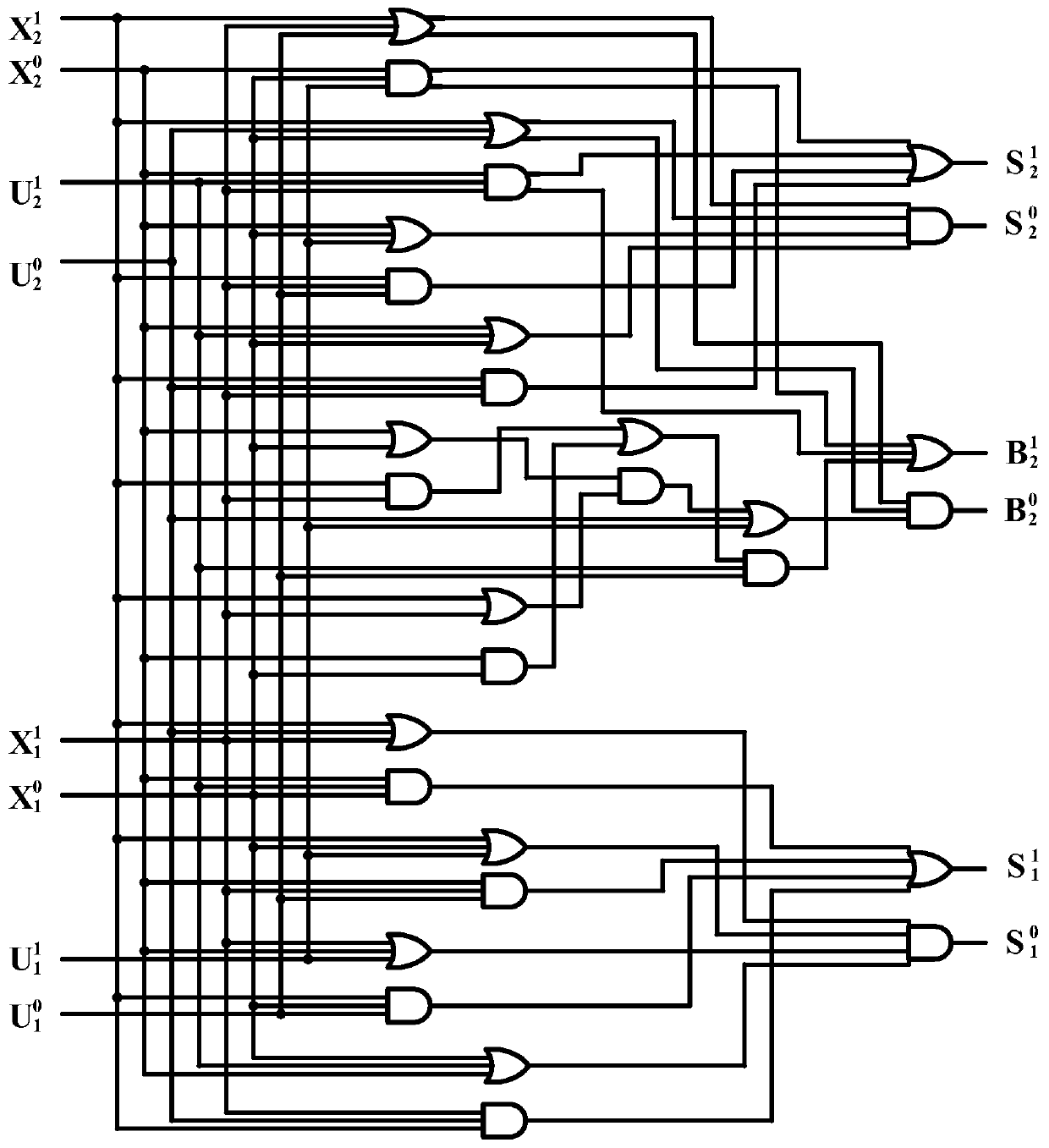
ActiveCN110532705AImprove propulsion performanceImprove reliabilitySpecial data processing applicationsDisplacement reactionsTruth table
The invention provides an implementation method of a two-bit Gray code subtractor molecular circuit based on DNA strand displacement. The implementation method is used for solving the problem of two-bit Gray code subtractor molecular operation based on Gray codes. The method comprises the following steps of: listing an operation truth table according to an operation rule of a two-bit Gray code subtractor circuit; converting the two-bit Gray code subtractor circuit into a two-bit Gray code subtractor double-track logic circuit by using a double-track idea; researching a DNA strand displacementreaction principle based on DNA strand displacement reaction; designing and cascading all DNA molecule fan-out doors, DNA molecule AND gates, DNA molecule OR gates and DNA report gate structures according to a DNA strand displacement reaction principle, and constructing a two-bit Gray code subtractor molecular circuit; visual DSD software is used for carrying out simulation analysis on the molecular circuit, and the dynamic characteristics of the molecular circuit are verified. According to the invention, a basic theoretical basis is provided for constructing a more complex logic operation circuit in the future, and the development of the biological computer is promoted, so that the reliability of the logic circuit of the biological computer is improved.
Owner:ZHENGZHOU UNIVERSITY OF LIGHT INDUSTRY
Logic circuit and device based on DNA strand displacement
ActiveCN111428877AHigh sensitivityImprove accuracyEnergy efficient computingDNA computersControl engineeringDna strand displacement
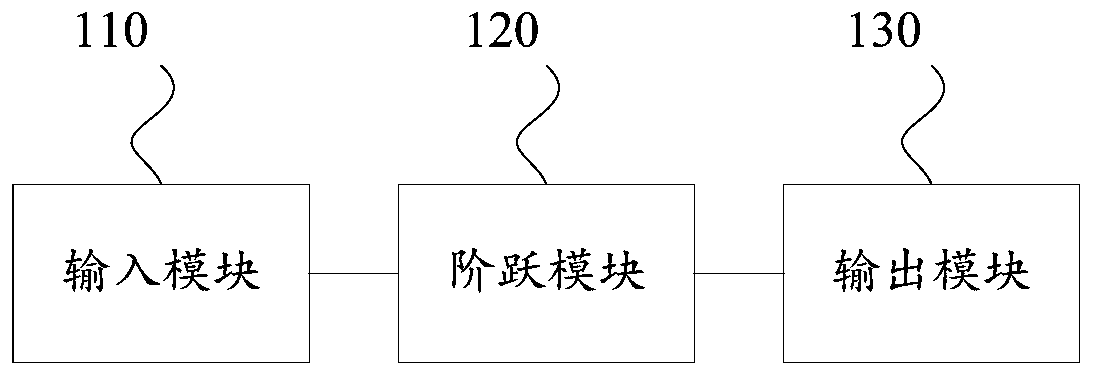
The invention discloses a logic circuit and device based on DNA strand displacement. The circuit comprises an input module used for obtaining an input signal single chain of a first concentration; thestep module is provided with a corresponding preset threshold value and a preset gain ratio, and is used for reacting to generate an output signal single chain with the concentration of the preset gain ratio when the total concentration of the input signal single chain is higher than the preset threshold value; and the output module is used for outputting a 0-1 digital signal according to the total concentration of the output signal single chain. The device comprises the circuit. According to the embodiment of the invention, the method comprises the steps of carrying out the processing, according to the invention, the step module is arranged in the logic circuit, the concentration of the output signal single chain is adjusted based on the corresponding preset threshold and preset gain ratio and the input signal single chain reaction, and the digital signal is finally output, so that the abrupt change sensitivity and accuracy of the circuit can be improved, and the working efficiency of the circuit formed based on the DNA chain can be improved. The method can be widely applied to the technical field of DNA information.
Owner:GUANGZHOU UNIVERSITY
Construction method of two-dimensional probability distribution model based on strand displacement
InactiveCN107480106AReduce calculationThe result is accurateDesign optimisation/simulationComplex mathematical operationsDna strand displacementTransducer
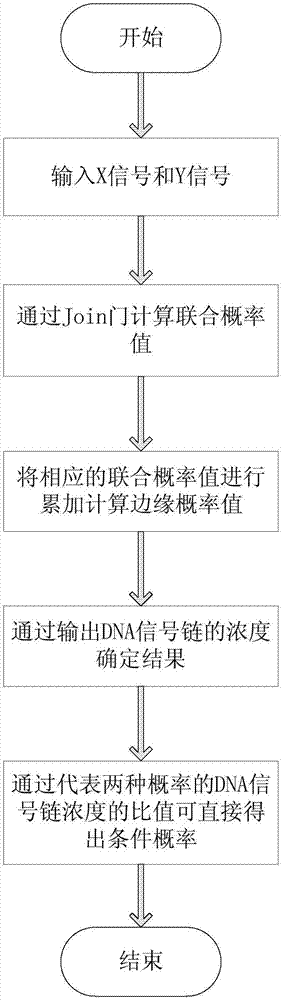
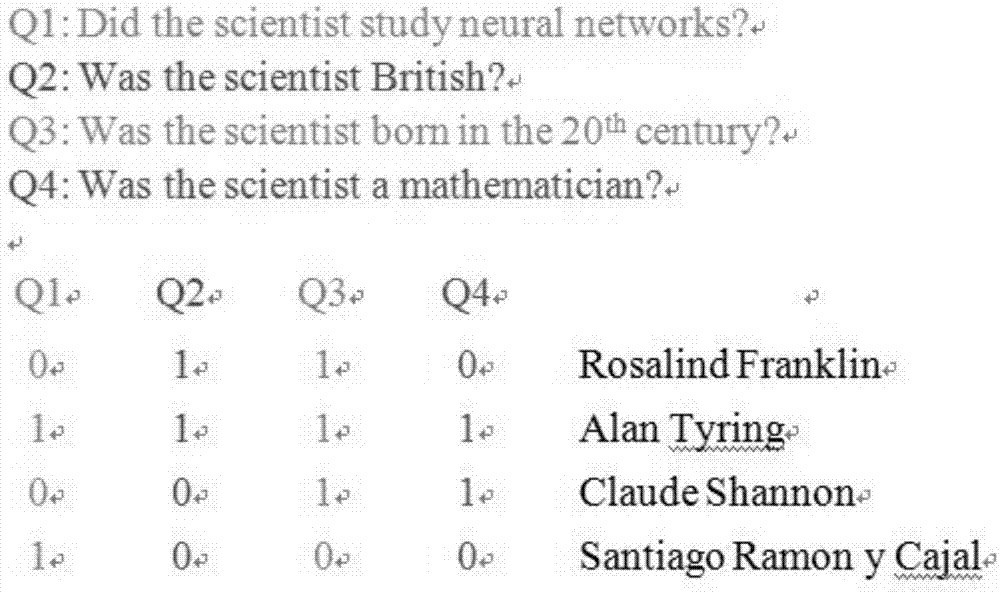
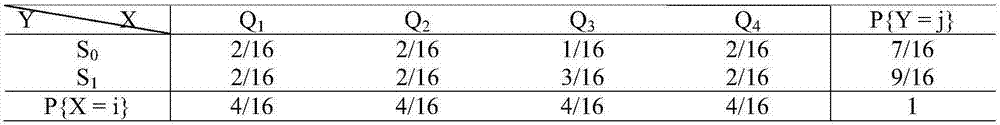
The invention relates to the field of strand displacement, and discloses a two-dimensional probability distribution model based on DNA strand displacement. Calculation of combined probability distribution, edge probability distribution and condition probability distribution is achieved. A join door module is utilized to calculate a combined probability distribution law, and a transducer door module is utilized to calculate an edge probability distribution law. A condition probability can be directly obtained through a combined probability and an edge probability. A specific case is utilized to conduct simulation verification on the model, as is proved by tests, the model can accurately calculate three two-dimensional probabilities, the error is small, the reaction speed is high, the result is stable, and the effectiveness of the two-dimensional probability distribution model is proved.
Owner:DALIAN UNIV
Biosensor based on strand displacement and dark-state silver clusters and application method of biosensor
ActiveCN108866157AImplement loop detectionIncreased sensitivityMicrobiological testing/measurementSilver clusterFluorescence
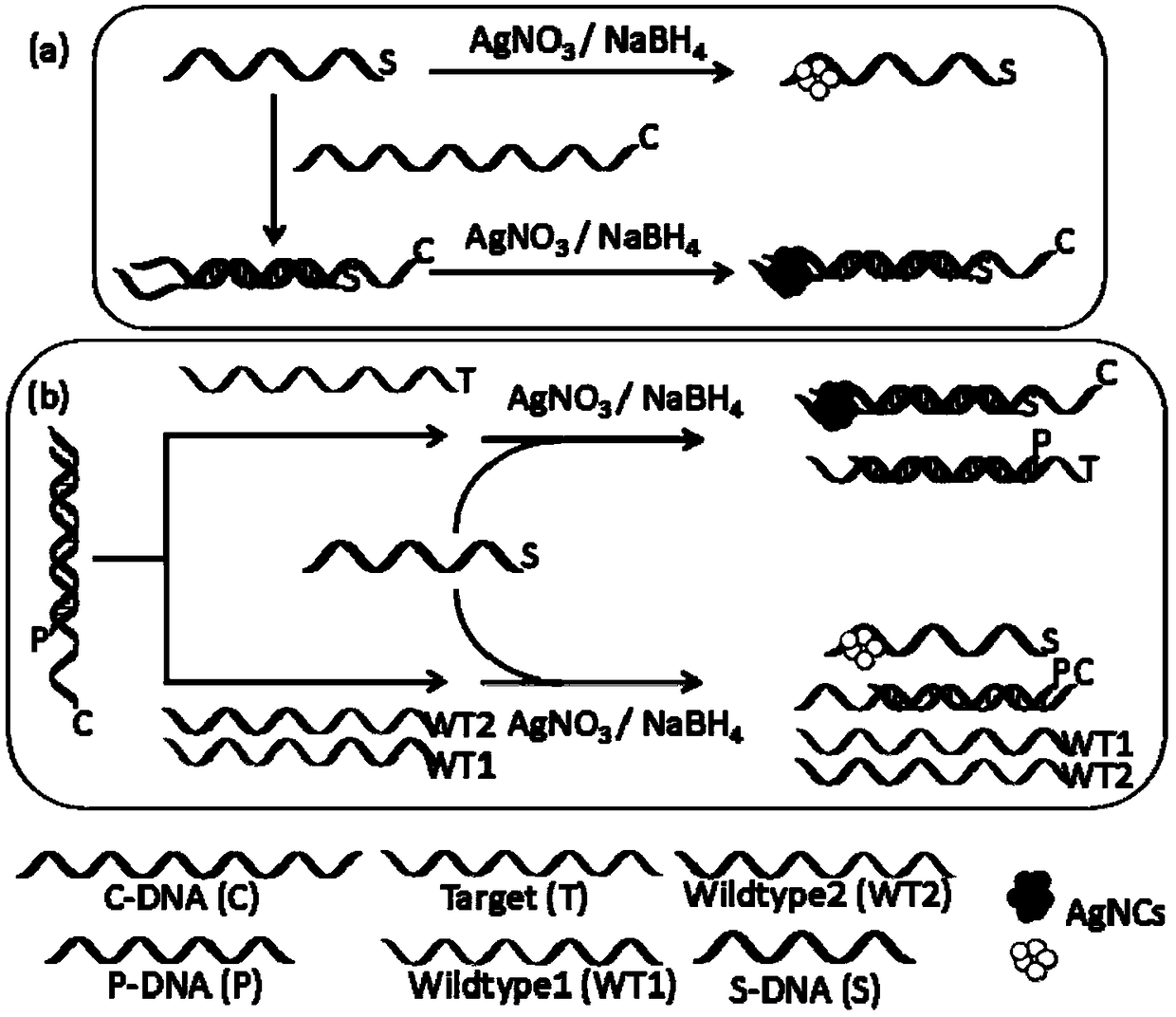
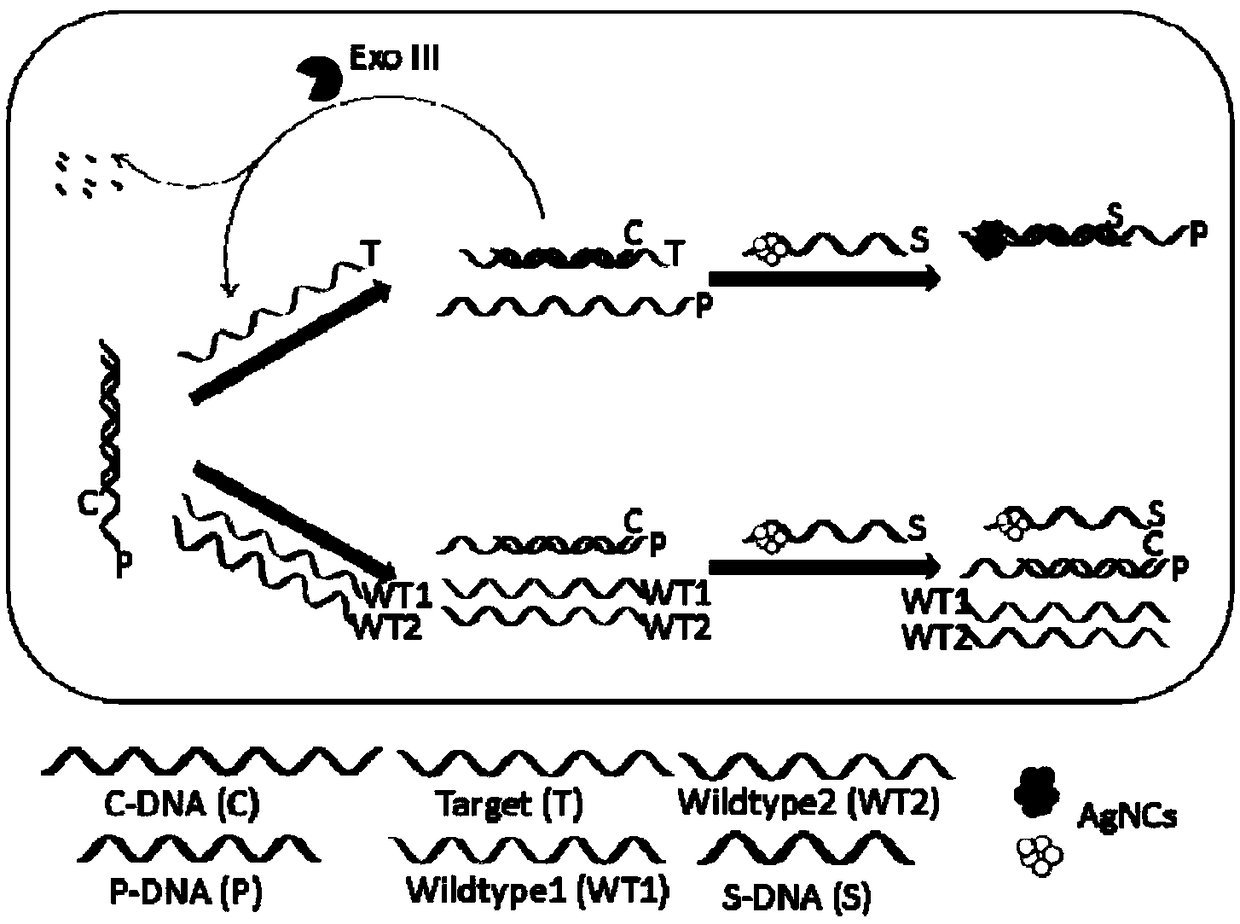
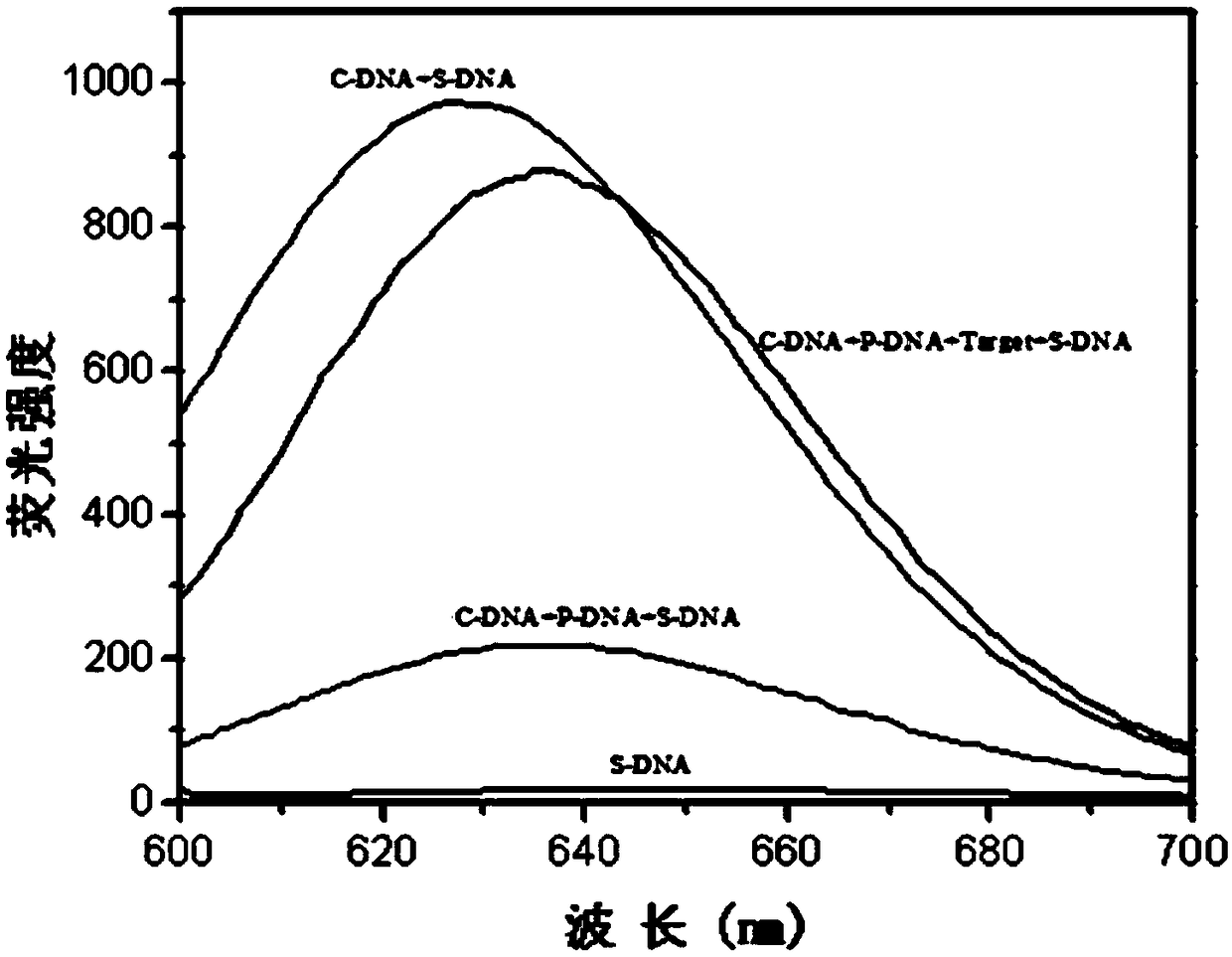
The invention relates to a biosensor based on strand displacement and dark-state silver clusters and application of the biosensor. The biosensor comprises three probe strands, including a C-DNA strand, a P-DNA strand and an S-DNA strand. The C-DNA strand comprises two components. The end 5' comprises a G-rich sequence. The end 3' complements the silver-cluster template strand S-DNA partially, andcomplements the probe strand P-DNA partially. The number of basic groups complementing to the P-DNA is larger than the number of basic groups complementing to the S-DNA. The P-DNA strand comprises a sequence capable of completely complementing to a target strand Target. The S-DNA strand comprises two components. The end 5' is a C-DNA complementing sequence, and the end 3' is a dark-state silver-cluster compounding template sequence. The biosensor and the application thereof have the advantages that an excision enzyme III is introduced into the system, the P-DNA in the Target / P-DNA double strand is cut, the Target is released to participate in P-DNA / C-DNA strand displacement, cyclic detection of the Target is realized, and detection sensitivity is improved; through fluorescence alteration of the dark-state silver clusters, strand displacement and amplification effect of the excision enzyme III, the biosensor can be applied to related gene detection such as sensitive gene deletion detection, and three base deletions can be distinguished obviously.
Owner:TONGJI UNIV
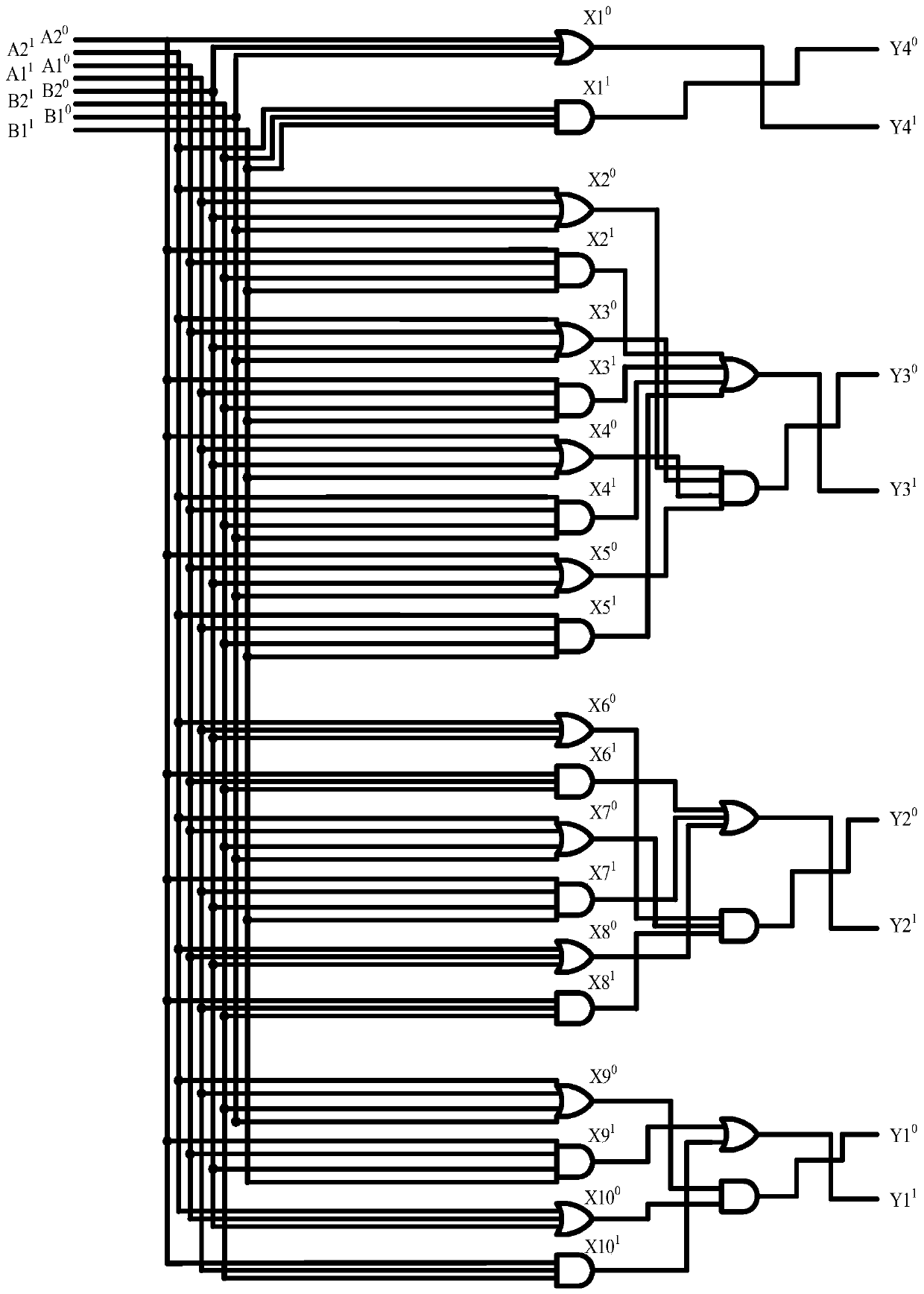
Mapping module-based 16-bit adder-subtractor design method
ActiveCN106126191AImprove certaintyAvoid instabilityComputation using non-contact making devicesLeft halfAdder–subtractor
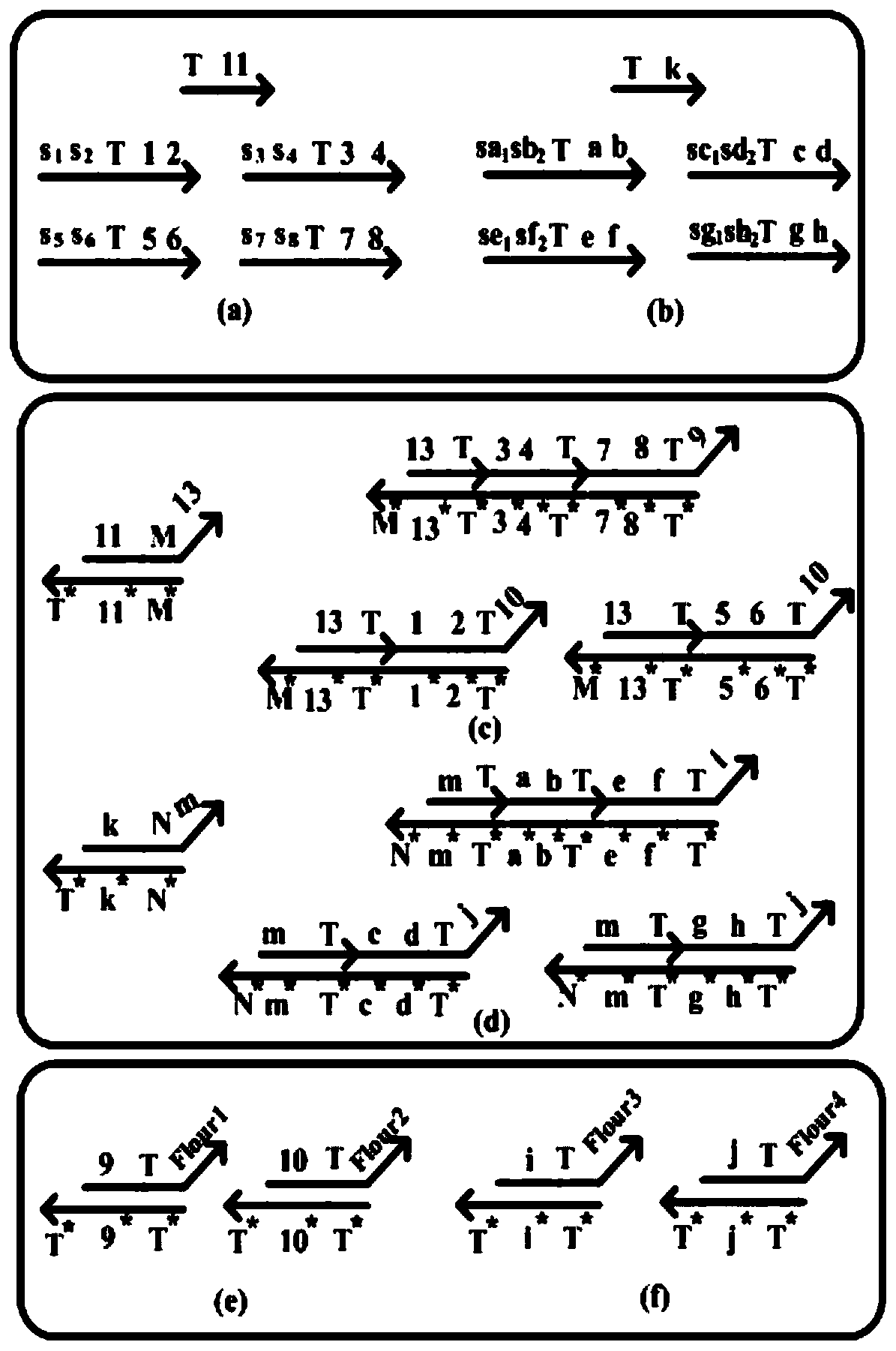
The invention relates to a method for designing a 16-bit adder-subtractor by utilizing mapping modules, and belongs to the technical field of computer application. The method comprises the following steps of 1: marking a DNA strand by utilizing a domain t and a domain f that represent logic 1 and logic 0 respectively, and establishing a domain-marked DNA signal strand that achieves a medium effect of transferring a signal in a DNA strand displacement reaction, wherein an input / output strand is the DNA signal strand, the DNA signal strand is symmetric in structure, the left half side of the DNA signal strand and a corresponding module of an upstream part can be subjected to the DNA strand displacement reaction, and the right half side of the DNA signal chain and a corresponding module of a downstream part can be subjected to the DNA strand displacement reaction; 2: establishing domain-marked 1 input-mapping modules; and 3: establishing domain-marked 2 input-mapping modules. The logic 1 and the logic 0 are represented through the domain t and the domain f respectively, so that the certainty of the reaction is enhanced and the instability caused by the fact that logic values are represented by concentration is avoided.
Owner:DALIAN UNIVERSITY
Odontoglossum ringspot virus RT-LAMP detection primer and detection method thereof
InactiveCN106367531AStrong specificityShorten the timeMicrobiological testing/measurementDNA/RNA fragmentationCymbidium mosaic virusFluorescence
The invention provides an odontoglossum ringspot virus RT-LAMP detection primer and a detection method thereof. The method comprises the following steps: designing primers by taking a CP gene sequence of cymbidium mosaic virus as a target gene, so as to obtain four specific primers in accordance with six regions; by virtue of a DNA strand displacement polymerase, conducting RT-LAMP amplification on the target gene with the application of the specific primers under a constant-temperature condition; and finally, conducting 1% agarose gel electrophoresis analysis, DNA fluorescent dye detection, enzyme restriction verification and cloning and sequencing contrasting on a reaction product. The loop-mediated isothermal amplification technique, which is established in accordance with odontoglossum ringspot virus, provided by the invention has the advantages of being rapid and efficient in amplification, good in specificity, simple and convenient to operate, free from the use of special instruments and the like; the technique has a relatively high application value and has a good application prospect in base and field detection.
Owner:CROP RES INST OF FUJIAN ACAD OF AGRI SCI
Method for detecting pine xylophilus, detection primers and lamp detection kit thereof
ActiveCN103555840BSimple and fast operationStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationFluorescenceDna strand displacement
The invention discloses a method for detecting bursaphelenchus xylophilus, a detection primer and a LAMP (loop-mediated isothermal amplification) detection kit of the detection primer. The method for detecting the bursaphelenchus xylophilus comprises the steps of (1) extracting a DNA (deoxyribonucleic acid) of the bursaphelenchus xylophilus in a pine wood sample; (2) constructing a LAMP reaction system for loop-mediated isothermal amplification; (3) judging whether the bursaphelenchus xylophilus exists in the pine wood sample according to the change of fluorescence or turdity of an amplified product. The invention also provides a method which is easy to operate, low in cost and high in stability and is used for extracting the DNA of the bursaphelenchus xylophilus from pine wood. The invention further provides a LAMP primer group with high specificity and high sensitivity for detection of the bursaphelenchus xylophilus. The invention also provides the LAMP detection kit for the bursaphelenchus xylophilus. The LAMP detection kit comprises the LAMP primer group, a reaction buffering solution, a DNA chain replacement enzyme and a fluorescent dye / specific probe. The detection kit disclosed by the invention has the advantages of high specificity, high sensitivity, convenience and quickness in detection and the like.
Owner:杭州益森键生物科技有限公司
Cymbidium mosaic virus RT-LAMP (reverse transcription-loop-mediated isothermal amplification) detection primers and detection method thereof
InactiveCN106191315AStrong specificityShorten the timeMicrobiological testing/measurementMicroorganism based processesCymbidium mosaic virusEnzyme digestion
The invention provides Cymbidium mosaic virus RT-LAMP (reverse transcription-loop-mediated isothermal amplification) detection primers and a detection method thereof. The primers are as follows: the nucleotide sequence of the outer primer F3 is disclosed as SEQ ID NO.2, the nucleotide sequence of the outer primer B3 is disclosed as SEQ ID NO.3, the nucleotide sequence of the inner primer FIP is disclosed as SEQ ID NO.4, and the nucleotide sequence of the inner primer BIP is disclosed as SEQ ID NO.5. The method comprises the following steps: designing primers by using CP gene sequence of Cymbidium mosaic virus as the target gene to obtain 4 specific primers for 6 regions, replacing the polymerase with a DNA chain, carrying out RT-LAMP on the target gene under homothermal conditions by using the specific primers, and finally, carrying out 1% agarose gel electrophoresis analysis, DNA fluorescent dye detection, enzyme digestion verification and clone sequencing comparison on the reaction product. The LAMP technique established according to the Cymbidium mosaic virus has the advantages of high amplification speed and efficiency, favorable specificity, no need of special apparatuses and the like, is simple to operate, has higher application value, and thus, has favorable application prospects in base layer and field detection.
Owner:CROP RES INST OF FUJIAN ACAD OF AGRI SCI
Low density parity check code decoding method based on chemical reaction network
ActiveCN108631791AReduce computing consumptionImprove parallelismError correction/detection using multiple parity bitsCode conversionChemical reactionDna strand displacement
The invention discloses a low density parity check code decoding method based on a chemical reaction network. The method performs probability number calculation by using a specific DNA strand concentration to represent a specific probability number and using a DNA strand displacement reaction. In the calculation process, several sets of DNA strand displacement reactions are pre-designed to performcalculations such as addition, subtraction, multiplication and division, and the respective group of reactions is sequentially controlled by a clock also composed of DNA strand displacement reactions. The invention is capable of implementing decoding calculations via pathways other than silicon.
Owner:SOUTHEAST UNIV
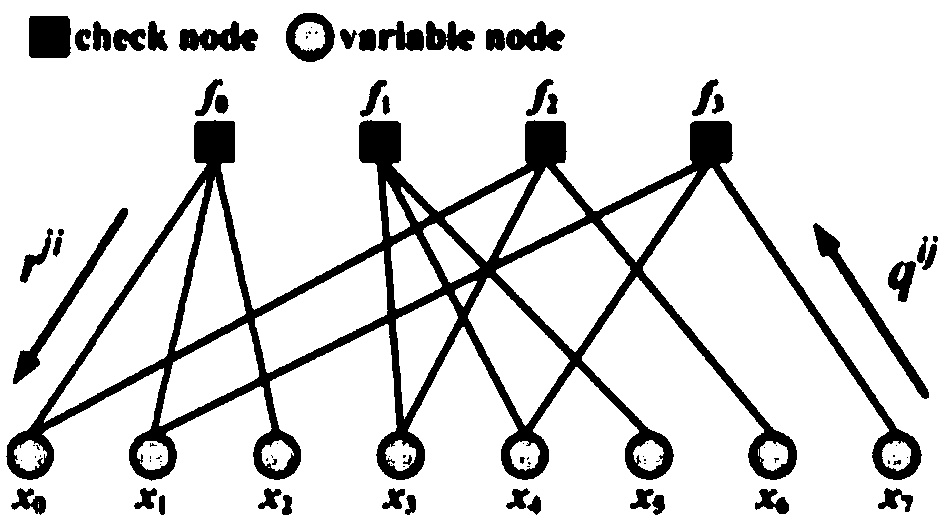
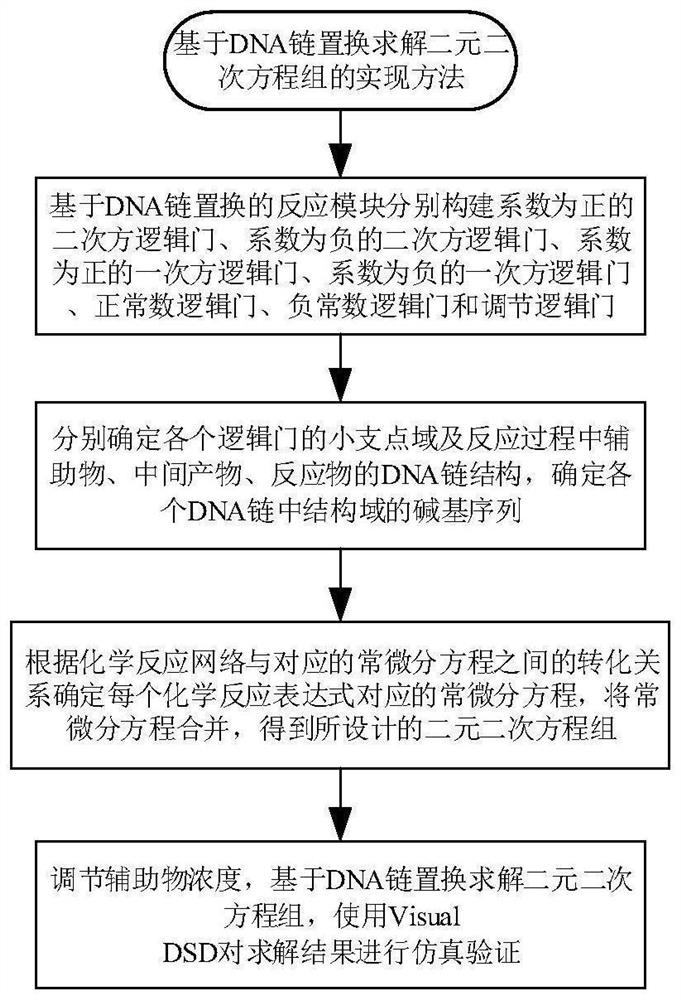
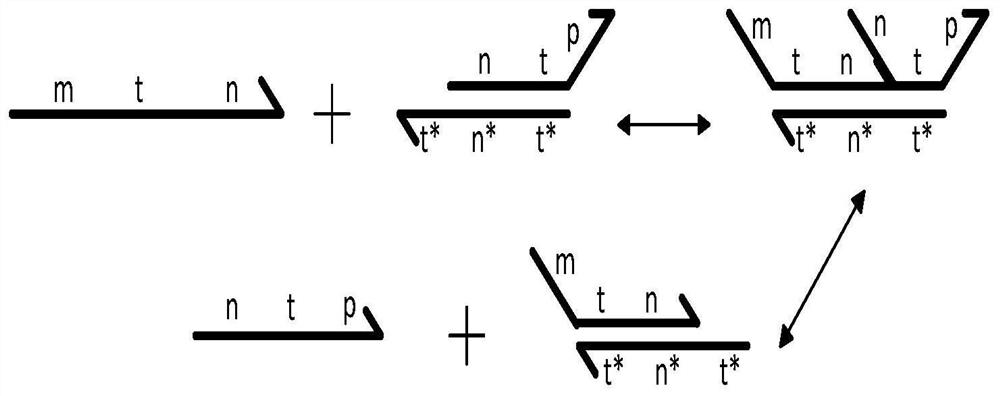
Implementation method for solving binary quadratic equation set based on DNA strand displacement
The invention provides an implementation method for solving a binary quadratic equation set based on DNA strand displacement. A reaction module based on DNA strand displacement respectively constructs a quadratic logic gate with a positive coefficient, a quadratic logic gate with a negative coefficient, a primary logic gate with a positive coefficient, a primary logic gate with a negative coefficient, a normal number logic gate, a negative constant logic gate and an adjusting logic gate; secondly, respectively a small fulcrum domain and a base sequence of the small fulcrum domain of each logic gate as well as DNA chain structures and base sequences of auxiliaries, intermediate products and reactants are determined respectively; a binary quadratic equation set is obtained according to the conversion relation between the chemical reaction network of each logic gate and the corresponding ordinary differential equation; and finally, the binary quadratic equation set is set by adjusting the concentration of the auxiliary, and simulation verification is carried out on a solving result by using Visual DSD. According to the method, solution of the binary quadratic equation set is realized based on the DNA strand displacement reaction, and the reasonability and effectiveness of the system are proved by utilizing a simulation result.
Owner:ZHENGZHOU UNIVERSITY OF LIGHT INDUSTRY
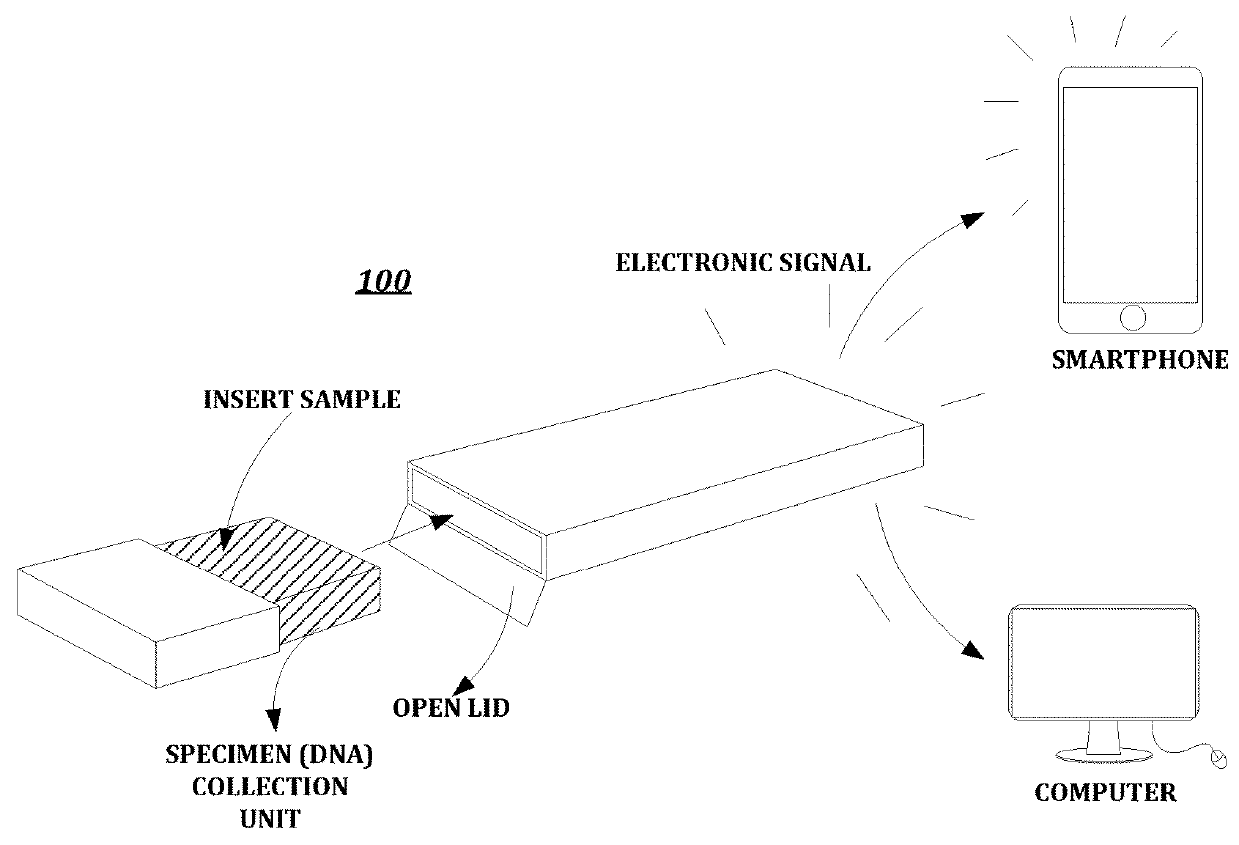
Kit capable of performing semiquantitative detection on micro ribonucleic acid 100 (microRNA100) directly
InactiveCN106834513AUndisturbedLow detection limitMicrobiological testing/measurementTotal rnaBiology
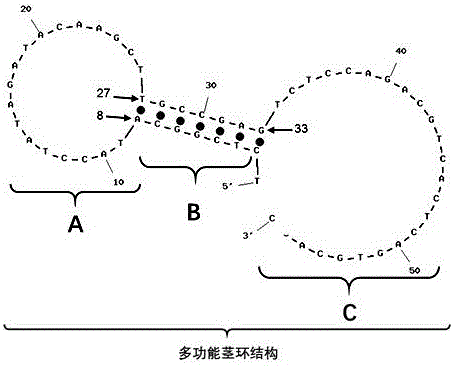
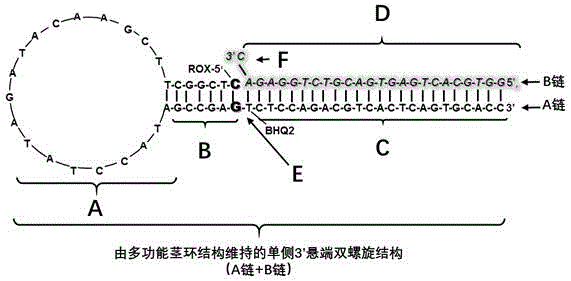
A kit capable of performing semiquantitative detection on micro ribonucleic acid (microRNA) directly is used for performing semiquantitative detection on certain microRNA in total RNA. The kit capable of performing semiquantitative detection on microRNA directly comprises a probe with a single-side 3' suspending end double helix structure maintained by a multifunctional stem-loop structure and a hybrid buffer solution containing DNA chain replacement enzyme; the probe with the single-side 3' suspending end double helix structure maintained by a multifunctional stem-loop structure is formed by assembling a chain A and a chain B which have different lengths and are oligonucleotide single chains; and the chain A is long and the chain B is short. The original sample, namely total microRNA extracting liquid, is directly added into a detection system, reverse transcription and amplification on a target sequence are avoided, and the kit is low in detection limit, short in detection time and applied in the technical fields of medicine and molecular biology.
Owner:中国医科大学
The Realization Method of the Molecular Circuit of Two-bit Gray Code Subtractor Based on DNA Strand Replacement
The invention provides an implementation method of a two-bit Gray code subtractor molecular circuit based on DNA strand displacement. The implementation method is used for solving the problem of two-bit Gray code subtractor molecular operation based on Gray codes. The method comprises the following steps of: listing an operation truth table according to an operation rule of a two-bit Gray code subtractor circuit; converting the two-bit Gray code subtractor circuit into a two-bit Gray code subtractor double-track logic circuit by using a double-track idea; researching a DNA strand displacementreaction principle based on DNA strand displacement reaction; designing and cascading all DNA molecule fan-out doors, DNA molecule AND gates, DNA molecule OR gates and DNA report gate structures according to a DNA strand displacement reaction principle, and constructing a two-bit Gray code subtractor molecular circuit; visual DSD software is used for carrying out simulation analysis on the molecular circuit, and the dynamic characteristics of the molecular circuit are verified. According to the invention, a basic theoretical basis is provided for constructing a more complex logic operation circuit in the future, and the development of the biological computer is promoted, so that the reliability of the logic circuit of the biological computer is improved.
Owner:ZHENGZHOU UNIVERSITY OF LIGHT INDUSTRY
A suture toehold activation method and toolkit for regulated DNA strand displacement reactions
The invention provides a suturing Toehold activation method used for controlling a DNA strand displacement reaction and a toolkit. A novel Toehold activation method is developed based on hairpin reconstruction caused by mutual action of a DNA hairpin structure and different environmental irritants (Hg2+ or ATP). The invention develops a novel Toehold activation method. The extra level of the DNA strand displacement reaction can be accurately controlled by changing the concentration of the environmental irritants.
Owner:SHANDONG UNIV
Method for detection of a nucleic acid target sequence
Disclosed is a method of facilitating detection of a nucleic acid target sequence. The method may include utilizing a toehold-mediated DNA strand displacement apparatus comprising a portion complementary to the nucleic acid target sequence. The method may further include utilizing a RNA toehold switch comprising a RNA sequence. Further, the toehold portion of the RNA sequence may be complementary to a portion of the toehold-mediated DNA strand displacement apparatus. The method may further include combining the toehold-mediated DNA strand displacement apparatus and the RNA toehold switch in an assay, such that the two are never in direct physical contact with each other. Accordingly, a sample containing the nucleic acid target sequence on the substrate may displace a nucleic acid strand from the toehold-mediated DNA strand displacement apparatus and bind a portion of it to the RNA toehold switch resulting in expression of the reporter protein.
Owner:TOTEMID INC
Molecular circuit design method based on four-input factorial addition operation based on DNA strand replacement
ActiveCN110544511BImprove reliabilityComputation using non-contact making devicesCAD circuit designSoftware engineeringDna strand displacement
Owner:ZHENGZHOU UNIVERSITY OF LIGHT INDUSTRY
Kit for directly performing semi-quantitative detection on microRNA675 (micro Ribonucleic Acid 675)
The invention discloses a kit for directly performing semi-quantitative detection on microRNA675 (micro Ribonucleic Acid 675), which is used for performing semi-quantitative detection on a certain microRNA675 (micro Ribonucleic Acid 675) in total RNA. The kit for performing the semi-quantitative detection on the microRNA675 (micro Ribonucleic Acid 675) comprises a probe with 'a single-side 3'suspension end double helix structure which is maintained by a multifunctional stem-loop structure' and a hybridization buffering solution which contains a DNA chain replacement enzyme; the probe with the 'single-side 3'suspension end double helix structure which is maintained by the multifunctional stem-loop structure' is formed by assembling an oligonucleotide single chain A and an oligonucleotide single chain B with different lengths; the chain A is longer, and the chain B is shorter. According to the kit for directly performing the semi-quantitative detection on microRNA675 (micro Ribonucleic Acid 675), an original sample, namely, a total microRNA675 (micro Ribonucleic Acid 675) extracting liquid, is directly added into a detecting system without performing reverse transcription and amplification on the microRNA675 (micro Ribonucleic Acid 675); the detecting limit is low; the detecting time is short; the kit is applied to the fields of medicines and molecular biology.
Owner:中国医科大学
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com