Kit capable of performing semiquantitative detection on micro ribonucleic acid 100 (microRNA100) directly
A microRNA, semi-quantitative technology, applied in the fields of medicine and molecular biology, can solve the problems of high price, low sensitivity, long time consumption, etc., and achieve the effect of short detection time, high specificity and low detection limit
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
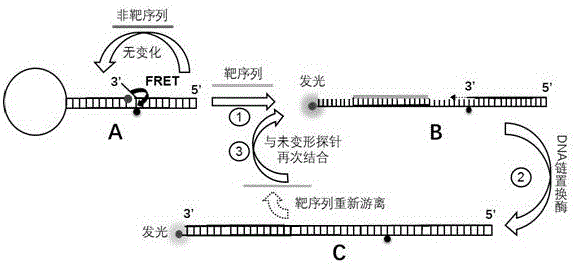
[0036] Using microRNA100-3p (hereinafter referred to as microRNA100) as the target sequence, semi-quantitative analysis was performed on different levels of microRNA100 standards.
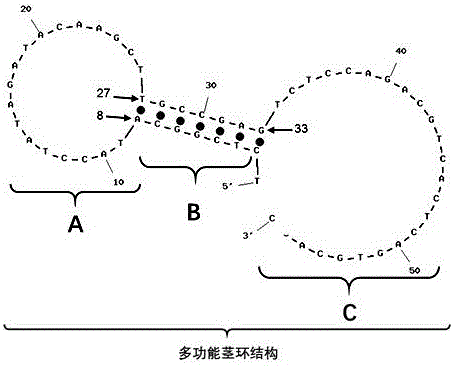
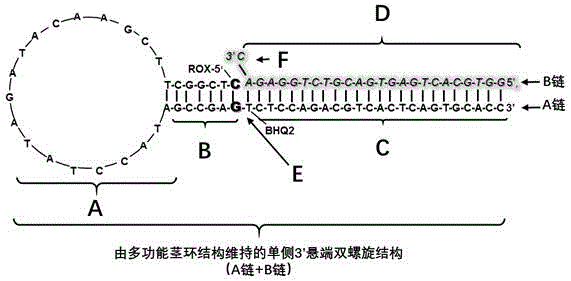
[0037] Step 1: Probe design and secondary structure prediction, the nucleotide sequence of microRNA100 is SEQ ID NO.3: 5'CAAGCTTGTATCTATAGGTATG 3', the nucleotide sequence and modification group of the A chain is SEQ ID NO.1: 5'ROXT C L TC L GGC ATACCTATAGATACAAGCTT GCCGAGT (-BHQ2)CTCCAGACGTCA
[0038] CTCAGTGCACC 3'. The single underlined sequence is complementary, the 5' end is modified with ROX (red fluorescent group), the double underlined T is modified with BHQ2 (fluorescent quencher group), the nucleotide with the upper right corner marked L is a locked nucleic acid, and the modification of the locked nucleic acid It is only used to increase the thermal stability of the probe's initial structure and reduce background fluorescence, and has no other effects. The prediction results of t...
Embodiment 2
[0046] The specificity of the probe was examined by using a mixture of high-concentration single-base mismatch sequences and double-stranded DNA identical to the microRNA100 sequence but twice its length as interfering substances.
[0047] Step 1: MicroRNA100 probe design and synthesis, see Step 1 and Step 2 of Example 1, the single-base mismatch nucleotide sequence is SEQ ID NO.4: 5'CAAGCTTGTAT T TATAGGTATG 3', compared with microRNA100 sequence, only one base T Differences. The sequence of the double-stranded DNA for interference is the DNA double helix structure formed by SEQ ID NO.5: 5'CAAGCTTGTATCTATAGGTATGCAAGCTTGTATCTATAGGTATG 3' and its complementary sequence.
[0048] Step 2: Take 1 μL of the probe with a concentration of 10 μmol / L and 18 μL of the hybridization buffer, a total of 19 μL, mix well and add to a 200 μL milky white eight-tube tube.
[0049] Step 3: Add 1 μL of microRNA100 standard substance to sample well A to make the content of microRNA100 in the samp...
Embodiment 3
[0053] The expression difference of microRNA100 in Hosepic cells and HIEC cells was detected.
[0054] Step 1: microRNA100 probe design and synthesis, see Step 1 and Step 2 of Example 1.
[0055] Step 2: Trizol method was used to extract total RNA from Hosepic cells and HIEC cells.
[0056] Step 3: Take 5 μL of the total RNA extracts from the above two samples and add them to 200 μL milky white eight-tube tubes.
[0057] Step 4: Add 1 μL of probe with a concentration of 10 μM and 14 μL of hybridization buffer, totaling 20 μL, mix well and send to qPCR instrument for detection.
[0058] Step 5: The detection condition is constant temperature at 60 ℃ for 30 minutes.
[0059] Step 6: Result analysis. Test results such as Figure 6 As shown, at 30 minutes, the fluorescence intensity of the sample containing the total RNA of Hosepic cells was about 8.0*10 6 , the fluorescence intensity of samples containing HIEC cell total RNA is about 7.0*10 6 Therefore, it is determined tha...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com