Method for implementing single-molecule chemical reaction network by DNA (deoxyribonucleic acid) strand displacement reaction
A chemical reaction network and DNA chain technology, applied in the field of single-molecule chemical reaction network, can solve the problem of high complexity and achieve the effect of reducing complexity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0019] The technical solution of the present invention will be further introduced below in combination with specific implementation methods and accompanying drawings.
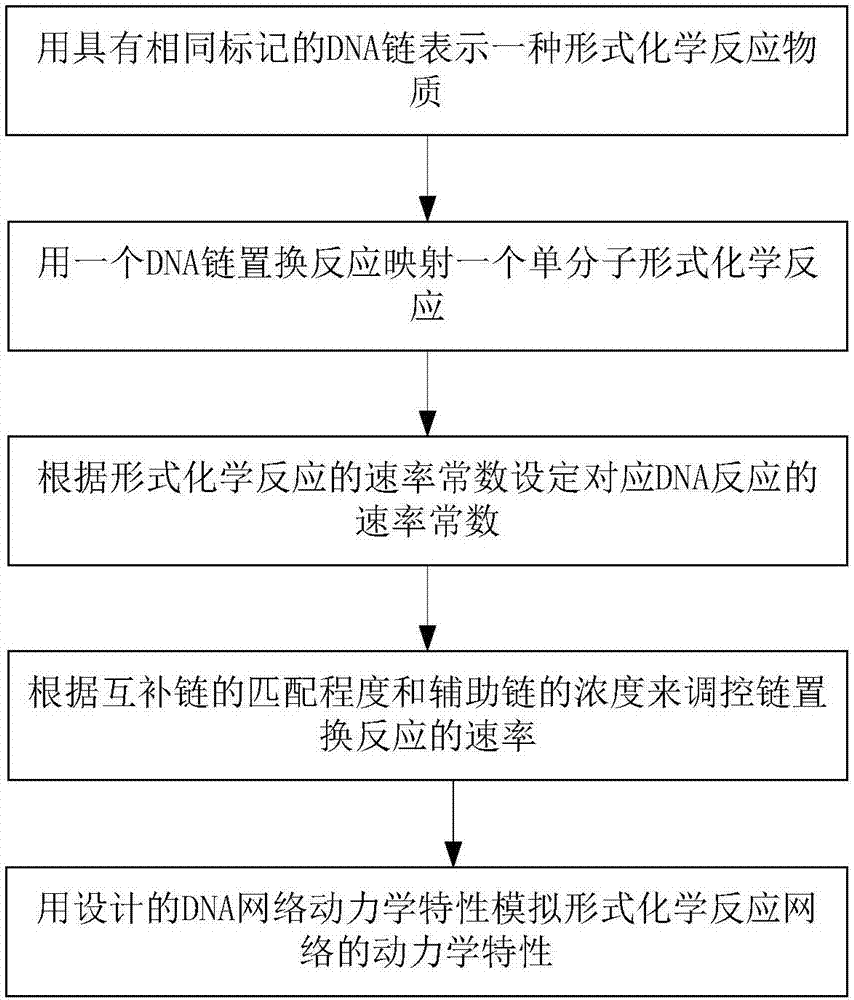
[0020] This specific embodiment discloses a method for realizing a chemical reaction network in the form of a single molecule by DNA strand displacement reaction, such as figure 1 shown, including the following steps:
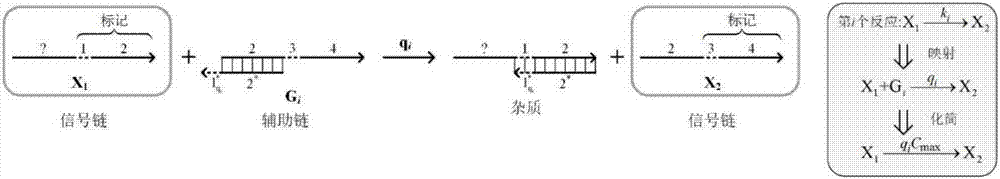
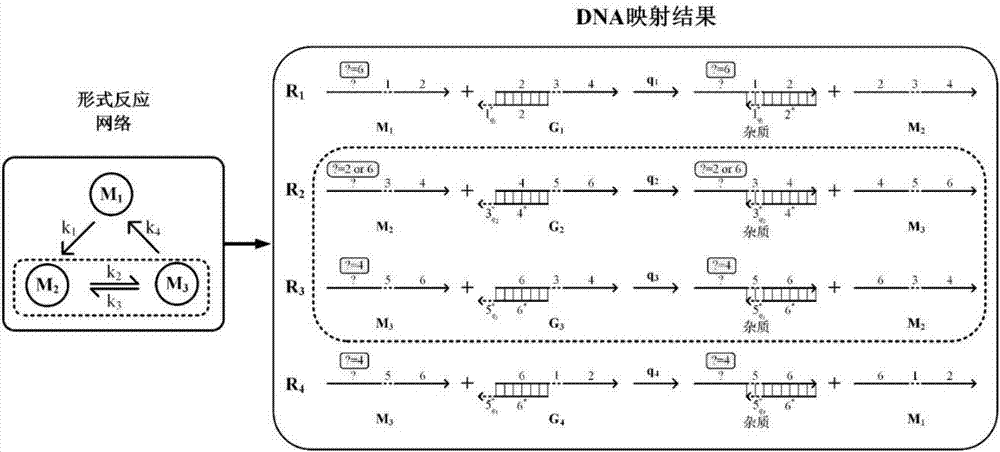
[0021] S1: Represent a form of chemical reaction substances with DNA strands with the same label, such as figure 2 As shown, each marker includes a short chain and a long chain, and each marker uniquely determines a form of reactant. All DNA strands with the same marker sequence, even if the sequence of the historical strand "?" Form reactants;
[0022] S2: if figure 2 As shown, a DNA strand displacement reaction is used to map an irreversible single-molecule chemical reaction, and two DNA strand displacement reactions are used to map a reversible single-molecule chemical reaction; the form...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com