Odontoglossum ringspot virus RT-LAMP detection primer and detection method thereof
A tooth blue ringspot virus, RT-LAMP technology, applied in biochemical equipment and methods, DNA / RNA fragments, recombinant DNA technology, etc., can solve the problems of high experimental equipment requirements, low sensitivity, unfavorable promotion, etc., to achieve Good application prospects, high application value, time-saving effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Dental ringspot virus RT-LAMP detection method, the steps are as follows:
[0038] Step 100: According to the relatively conservative CP gene sequence of ORSV published by GenBank, use the LAMP primer design software primer explorer 4.0 (http: / / primerexplorer.jp / elamp4.0.0 / index.html) to design primers for the target gene, and obtain 6 4 specific primers for the region. The 4 specific primer pairs are specifically:
[0039] Outer primer F3: 5'-TGTCTTACACTATTACAGACCCG-3';
[0040] Outer primer B3: 5'-TCATAGCGATAAACTCTGAAGT-3';
[0041] Internal primer FIP: 5'-GGAACTGATTACCCAGAGAATTGGTA-gaattc-TTATTTAAGCTCGGCTTGGGCTG-3';
[0042] Internal primer BIP: 5'-CTGTTCAACAGCAGTTTGCTGATG-gaattc-GCAGGGAACCTACTGGTCAAAG-3'.
[0043] The CP gene sequence of the blue ringspot virus is shown in SEQ ID NO.1; the nucleotide sequence of the outer primer F3 is shown in SEQ ID NO.2, and the nucleotide sequence of the outer primer B3 is shown in Shown in SEQ ID NO.3; the nucleotide sequen...
Embodiment 2
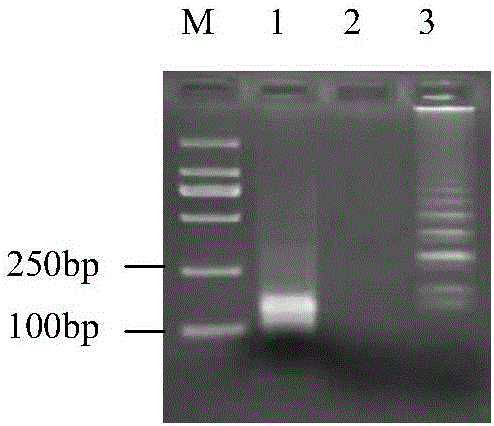
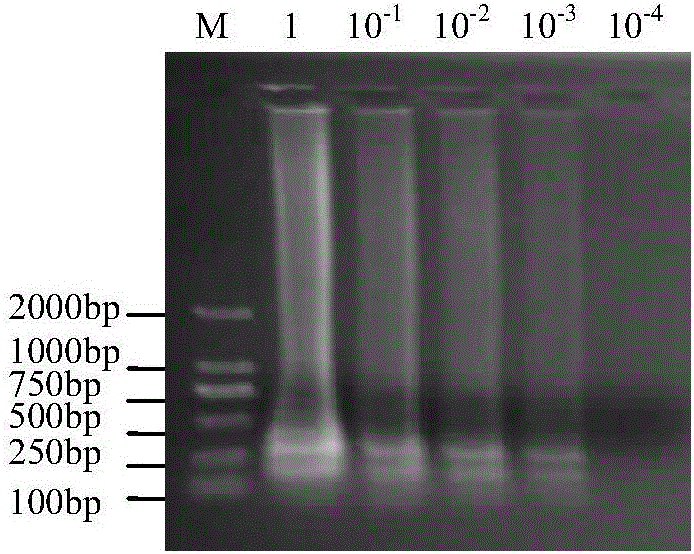
[0053] In order to confirm the correct amplification of the LAMP reaction, EcoRI restriction sites were introduced into FIP and BIP when designing LAMP primers in this experiment. After the LAMP reaction product in step 400 of Example 1 was digested with EcoRI enzyme at 37°C overnight, the digested product was run on the gel, and the electrophoresis results were as follows: figure 2 As shown, each lane is indicated as follows: M: DL2000 DNA marker, 1: restriction product, 2: negative control, 3: LAMP product. figure 2 It indicated that the ladder-like band amplified by LAMP could be successfully digested by EcoRI, and the size of the digested product was similar to the expected size.
[0054] In order to further verify the correctness of the LAMP product, the digested product was gel recovered, connected to PMD18-T to construct a recombinant plasmid containing the target fragment, transformed with JOM109 competent cells, and the transformed bacteria were sent for testing.
...
Embodiment 3
[0058] Specificity verification of RT-LAMP:
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com