Local DNA hairpin strand displacement reaction-based XOR gate and complementing circuit
A technology of chain displacement reaction and inversion circuit, which is applied in CAD circuit design, electrical digital data processing, special data processing applications, etc. It can solve the problems of slow molecular diffusion and wrong molecular collision, and achieve fast construction speed and good performance. , the effect of improving the accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
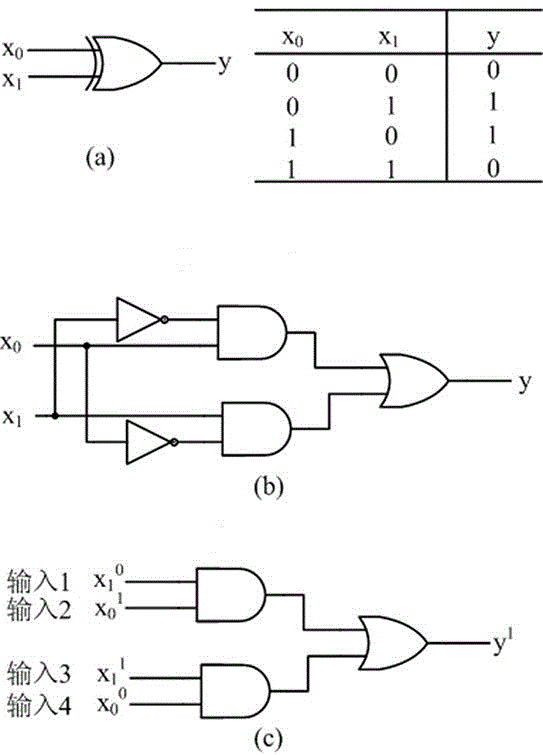
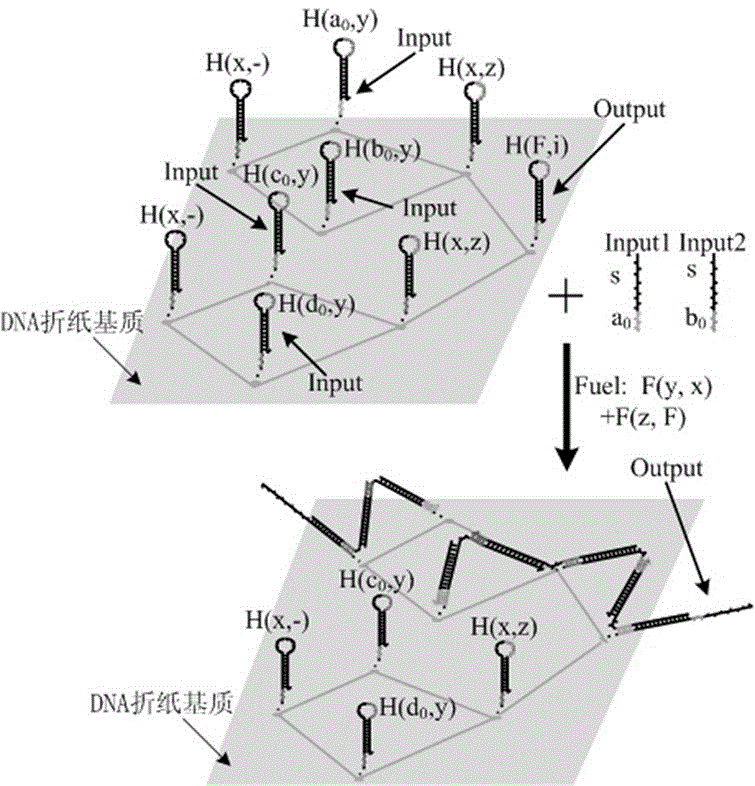
[0030] like figure 2As shown, the XOR gate based on the local DNA hairpin strand displacement reaction provided by the present invention is constructed by dual-track logic, including single-stranded DNA input, fuel hairpin and DNA hairpin fixed on the origami substrate;
[0031] The single-stranded DNA input has four: 0 >, 0 >, 0 >, 0 >
[0032] There are two kinds of DNA fuel cards: [s*]{x^> (that is, F(y, x) in the figure), [s*]{F^> (that is, in F(z, F);
[0033] There are seven types of DNA hairpins fixed on the origami substrate: {tether() a 0 ^*}[s]{y^> (that is, H in the figure (a 0 , y), {tether() x^*}[s]{blank^> (that is, H(x, -) in the figure, {tether() b 0 ^*}[s]{y^> (that is, H in the figure (b 0 , y), {tether() x^*}[s]{z^> (that is, H(x, z) in the figure, {tether() c 0 ^*}[s]{y^> (that is, H in the figure (c 0 , y), {tether() d 0 ^*}[s]{y^> (that is, H(d in the figure 0 , y), {tether() F^*}[s]{i^> (that is, H(F, i) in the figure, where tether() means that...
Embodiment 2
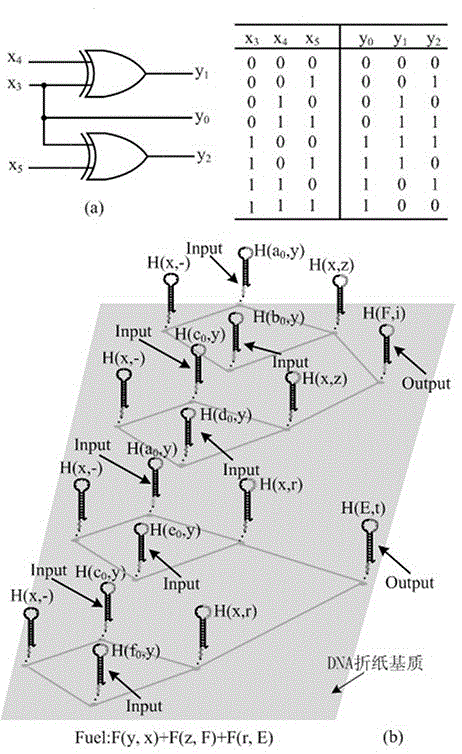
[0289] like image 3 As shown, the negation circuit constructed based on the local DNA hairpin displacement reaction XOR gate described in Example 1 belongs to a three-bit binary input negation circuit, which is composed of two parallel XOR gates;
[0290] In this circuit: input x 3 Represents the sign bit of the input binary number, it only has two logic states of "0" and "1", and "0" means that the input binary number is positive, "1" means that the input binary number is negative; input x 4 and x 5 Represent the high and low bits of the binary input value respectively, and also only have two logic states of "0" and "1"; output y 1 and y 2 Respectively represent the high and low bits of the binary output value, and also only have two logic states of "0" and "1";
[0291] The truth table for this negation circuit is as follows:
[0292] .
[0293] It can be seen from the truth table of the negation circuit that when the sign bit x of the binary number is input 3 When...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com