Elisa detection chip based on nucleic acid sequence coding and its preparation and application
A technology for detecting chips and nucleic acid sequences, which can be used in measurement devices, laboratory containers, chemical instruments and methods, etc., and can solve the problems of low detection throughput, poor ease of operation, and small number of codes.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
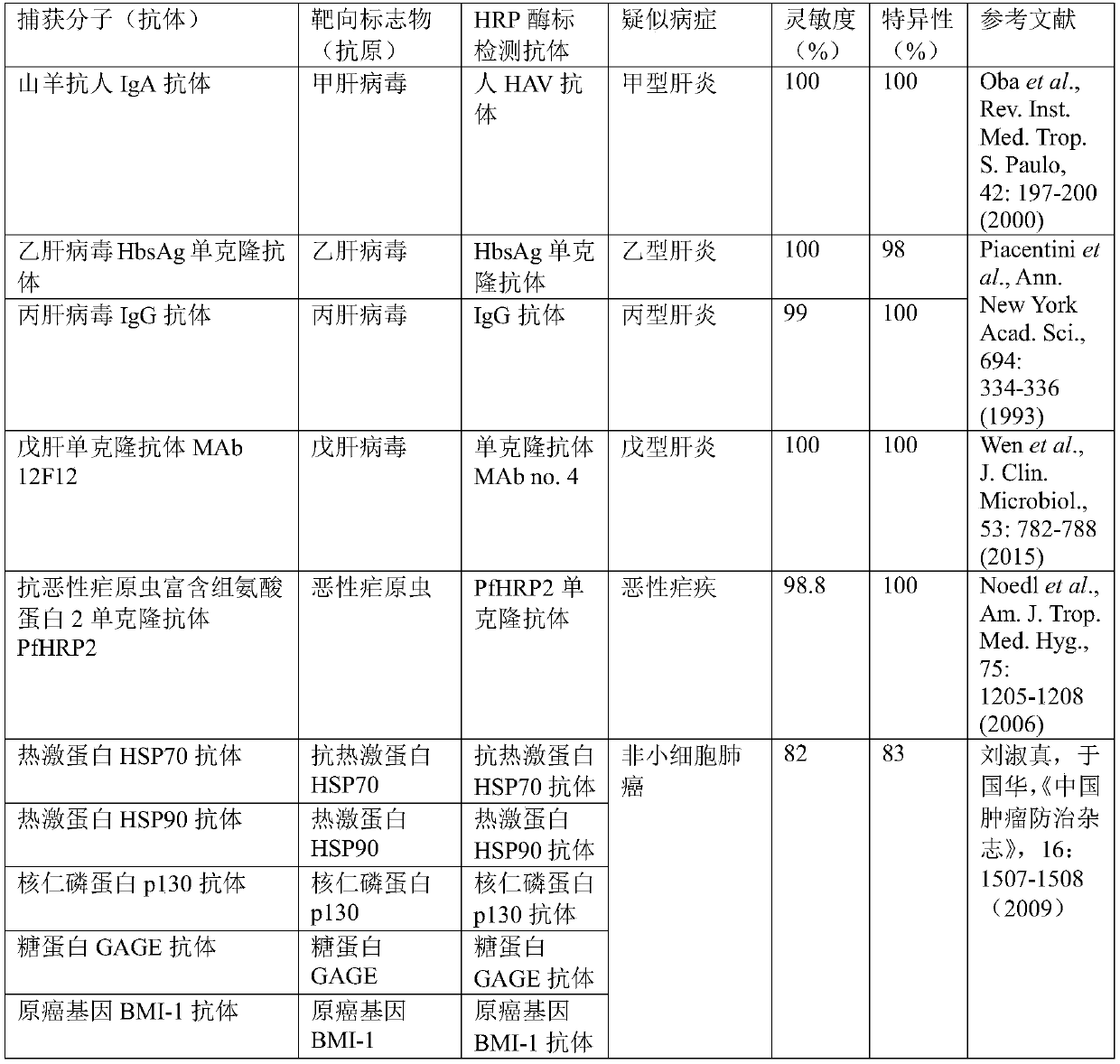
[0067] This embodiment provides a preparation and application based on an ELISA chip, specifically related to the double-antibody sandwich method, multi-person, multi-marker detection, as follows:
[0068] Step 1, surface modification of micron magnetic beads: The surface of about 1.1 million magnetic beads with a diameter of 3.5 μm is coated with anti-streptavidin (SA for short) by biochemical methods, and the density of SA on the surface of the magnetic beads is about 4000 / μm 2 , Therefore, the total amount of the surface area SA of a magnetic bead with a diameter of 3.5 μm is about the product of its surface area S and density, that is, 4000 beads / μm 2 ×4πr 2 ≈150,000 SAs. Divide the magnetic beads into 10020 equal parts, each about 110 beads; repeat three times.
[0069] Step 2, microbead coding: take the first 10020 sequences from the primary coding library, design and synthesize the hairpin DNA with the coding sequence, and the top of the loop of the hairpin structure ...
Embodiment 2
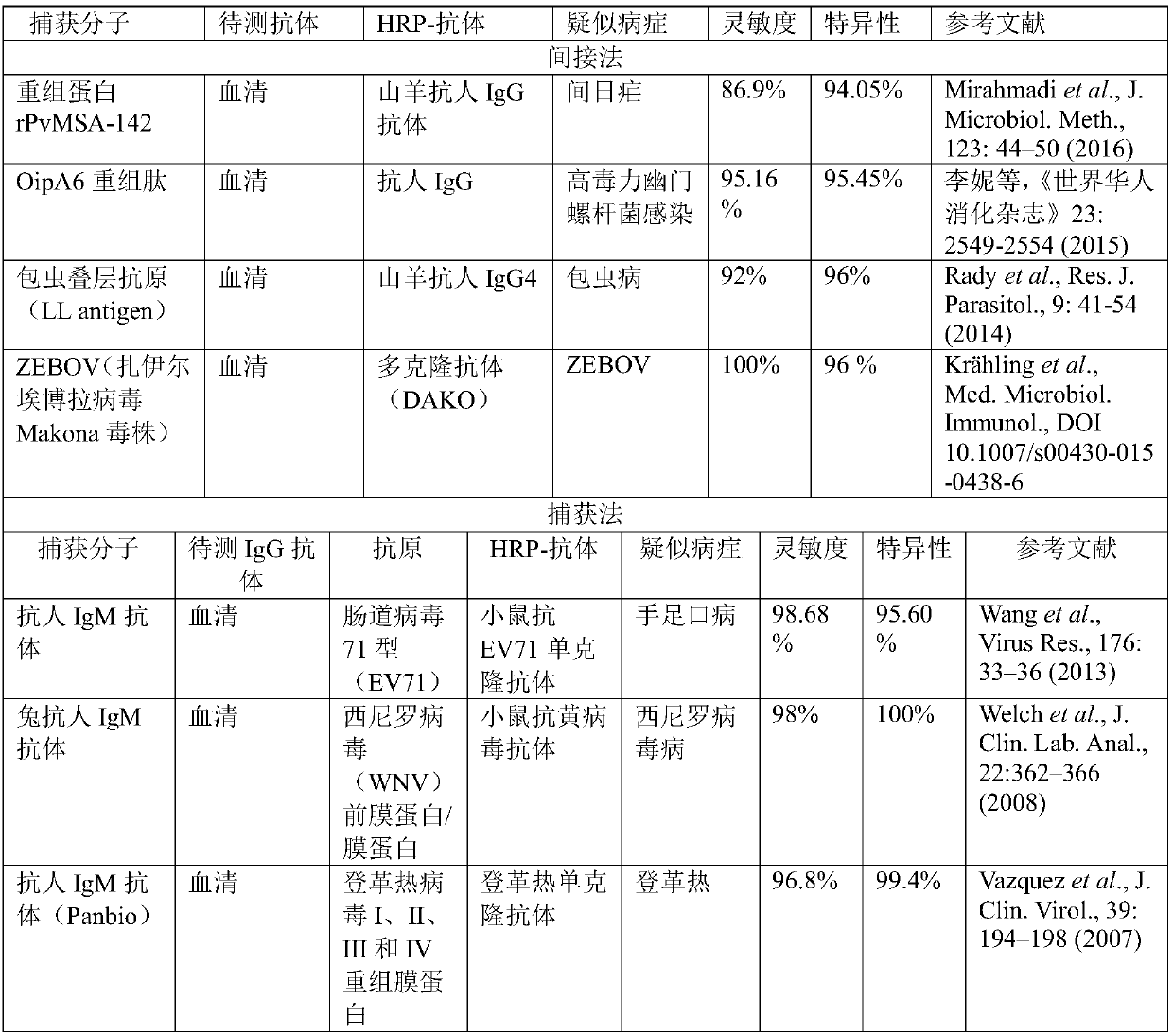
[0080] This embodiment provides a preparation and application based on an ELISA chip, involving indirect method: known antigen-test antibody-detection secondary antibody, capture method: IgM anti-antibody-test IgM antibody-specific antigen-labeled antibody, single The comprehensive detection of markers in multiple populations is as follows:
[0081] Step 1, surface activation of polystyrene (PS) micro-beads: use chemical methods to coat the surface of about 710,000 PS beads with a diameter of 2 μm with glutaraldehyde, divide the beads into 7 groups, each group has 1002 parts, and each part has about 100 capsules; repeat three times.
[0082] Step 2, PS bead coding and capture molecule immobilization: Synthesize amino-modified coding DNA, which consists of two parts, with an amino-labeled primer pairing region at the 3' end, sequence 3'NH 2 -CAGCACTGACCCTTTTGGGACCGC-5', followed by the coding region, which was taken from the first 7014 sequences of the preferred coding library...
Embodiment 3
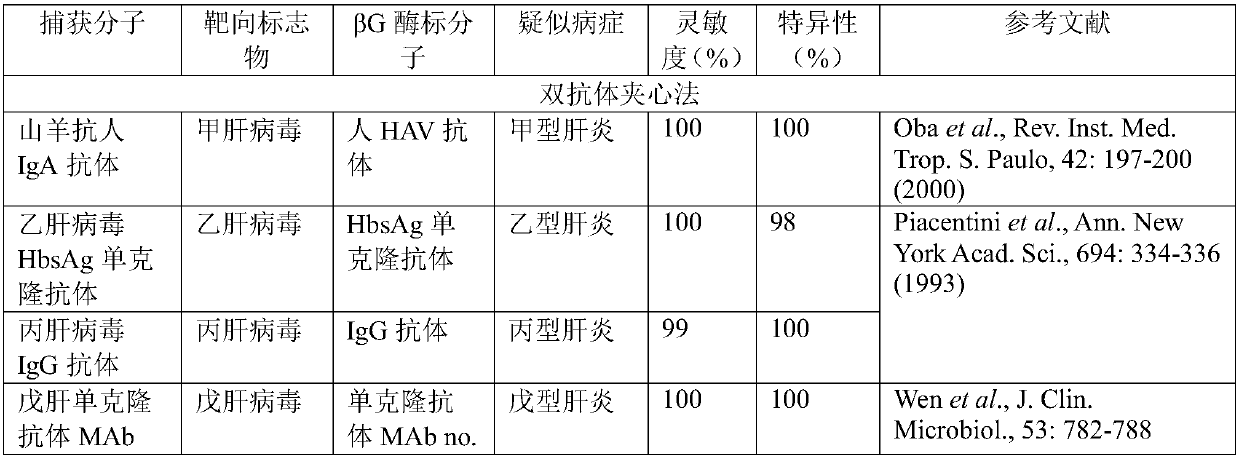
[0093] This example provides a preparation and application based on an ELISA chip, involving double-antibody sandwich method, double-antigen sandwich method and indirect method, multi-person multi-marker detection, details are as follows:
[0094] Step 1, surface activation of silicon nano-beads: Divide about 15 million silicon nano-beads with a diameter of 350 nm activated by SA on the surface into 36 groups, of which 12 groups of test beads each have about 1.2 million beads, and the other 24 groups of control beads have about 500 beads each , that is, 12 groups of positive and negative controls. Repeat 2 times.
[0095] Step 2, immobilization and DNA encoding of capture molecules on silica beads: 12 kinds of biotin-labeled capture molecules (Table 3) were mixed with the nanobeads used in each group for detection, positive control and negative control group, and immobilized on the surface of the nanobeads. The number of capture molecules is equivalent to about 1 / 3 of the num...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com