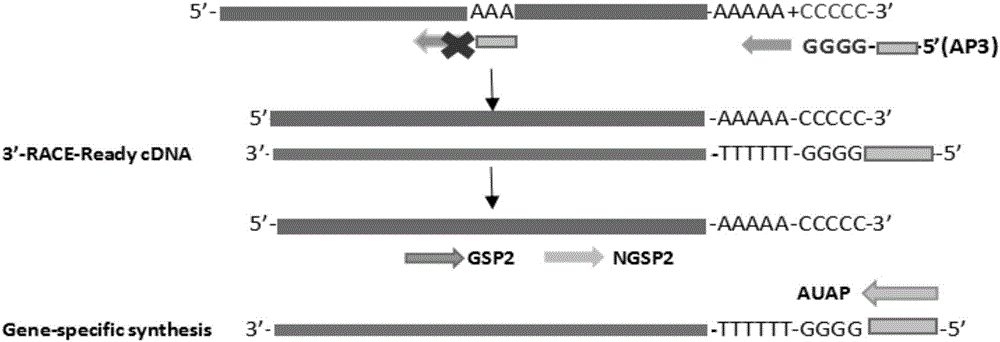
3'RACE (rapid amplification of cDNA ends) method for acquiring complete 3' end sequence as 3' end having repetitive sequence
A technology of repeated sequences and terminal sequences, applied in the biological field, can solve the problem of inability to obtain complete terminal fragments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] The mangrove plant kandelia (Kandelia obovata) iron-type superoxide dismutase (Fe-Superoxide Dismutase) gene was used as the test object, named as FeSOD1. After the total RNA of candela was extracted and passed the quality inspection, it was stored at ultra-low temperature (-80°C) for future use.
Embodiment 2
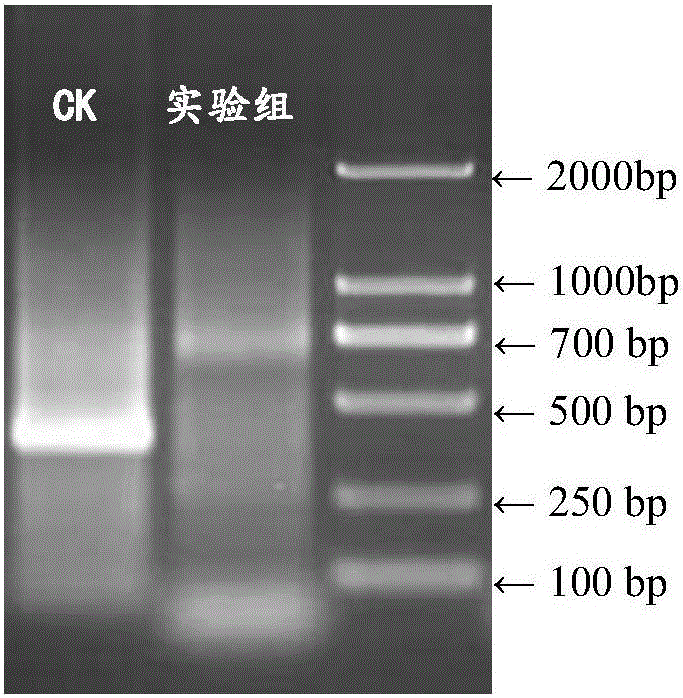
[0022] Control group: cDNA was reverse transcribed using Thermo Scientific RevertAid First Strand cDNASynthesis Kit (Fermentas EU) according to the general method.
Embodiment 3
[0024] Experimental group: (1) mRNA was tailed with terminal transferase from TaKaRa Company.
[0025] Prepare the following reaction solution in a microcentrifuge tube, the total volume is 50ul (Table 1).
[0026] Table 1 Preparation of TdT reaction solution
[0027]
[0028] Mix the reaction solution evenly, shake slightly and react at 37°C for 30 minutes. React at 80°C for 30 seconds to terminate the reaction.
[0029] (2) Reverse transcription experiments were performed using the Thermo Scientific RevertAid First Strand cDNA Synthesis Kit (Fermentas EU) kit. The preparation of the specific reaction solution is shown in Table 2.
[0030] Prepare the following reactions (Table 2) in microcentrifuge tubes.
[0031] Table 2 Preparation of reaction solution
[0032]
[0033] Note: AP3-GGCCACGCGTCGACTAGTACGGGGGGGGGGGGGGGGGG
[0034] React at 65°C for 5 minutes to open the RNA secondary structure and facilitate primer binding. React on ice for 1 minute.
[0035] ④ P...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com