Screening and application of biomarkers related to severe oligoasthenospermia
A biomarker, asthenozoospermia technology, used in instruments, measuring devices, scientific instruments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0124] Example 1: Screening of biomarkers associated with severe oligoasthenospermia
[0125] The specific screening method is as follows:
[0126] 1. Sample processing and experimental analysis
[0127] 1. Extraction of the whole protein of sperm cells: the same amount of severe oligospermia and normal sperm samples were washed three times with DPBS, and the same amount of RIPA lysate was added to sonicate for 1-2 minutes, incubated on ice for 30 minutes to lyse, and centrifuged at 4°C at 14,000g ×20min to take the supernatant. Protein concentration was determined using the Bradford method.
[0128]2. Proteolysis: Take approximately 150 μg of sperm protein from the samples of severe oligospermia and normal sperm, use 10% polyacrylamide gel electrophoresis (SDS-PAGE) to separate the proteins, and divide each into 5 parts for enzymatic hydrolysis. Peptides were desalted using ziptip.
[0129] 3. Mass Spectrometry: Nanoflow Liquid Chromatography Separation: Phase A: Water co...
Embodiment 2
[0185] Embodiment 2: clinical detection verification
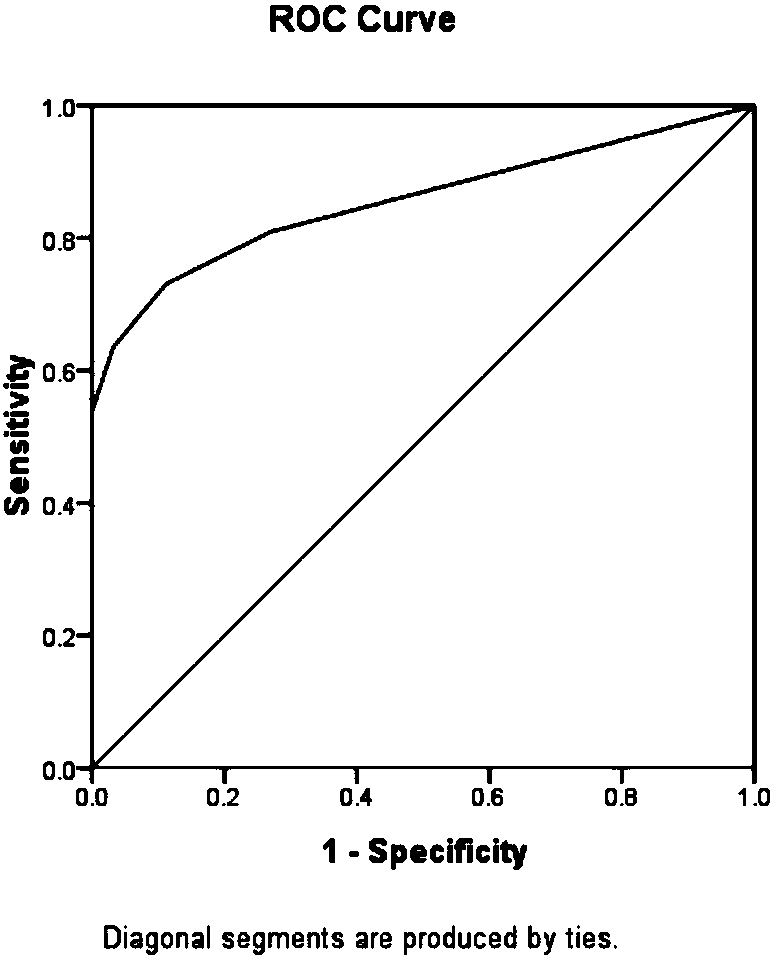
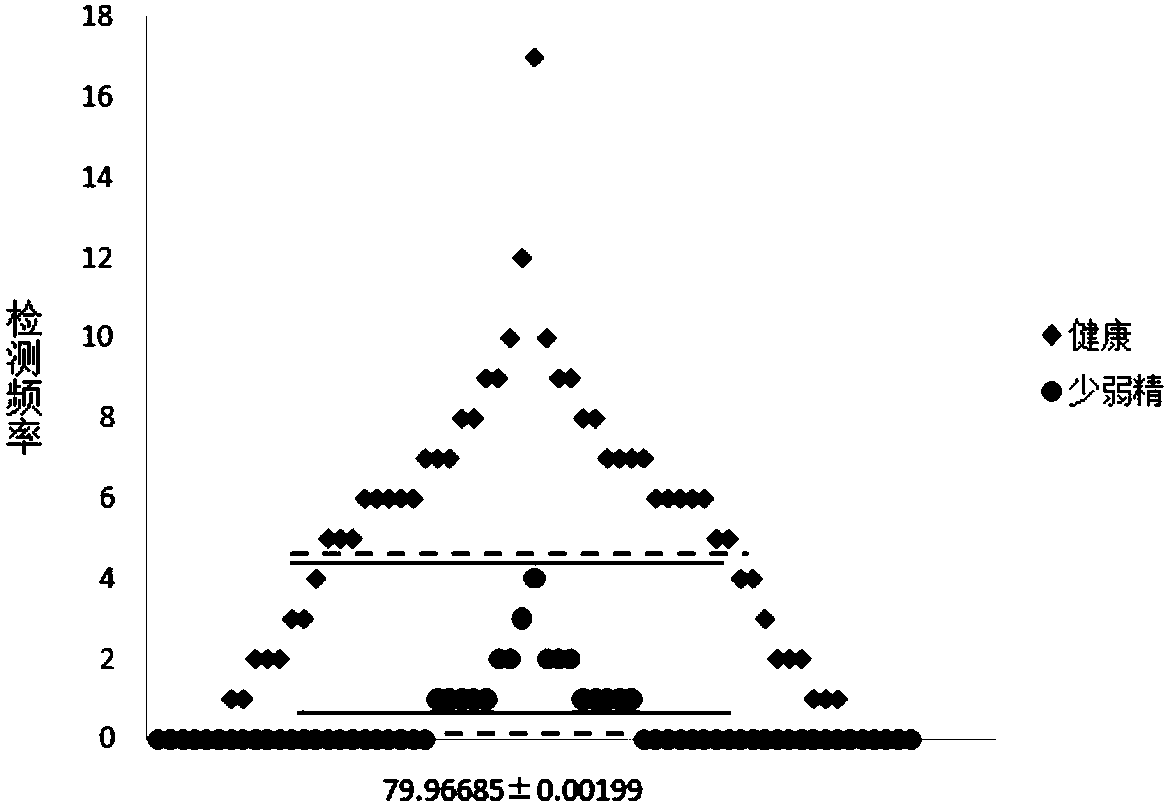
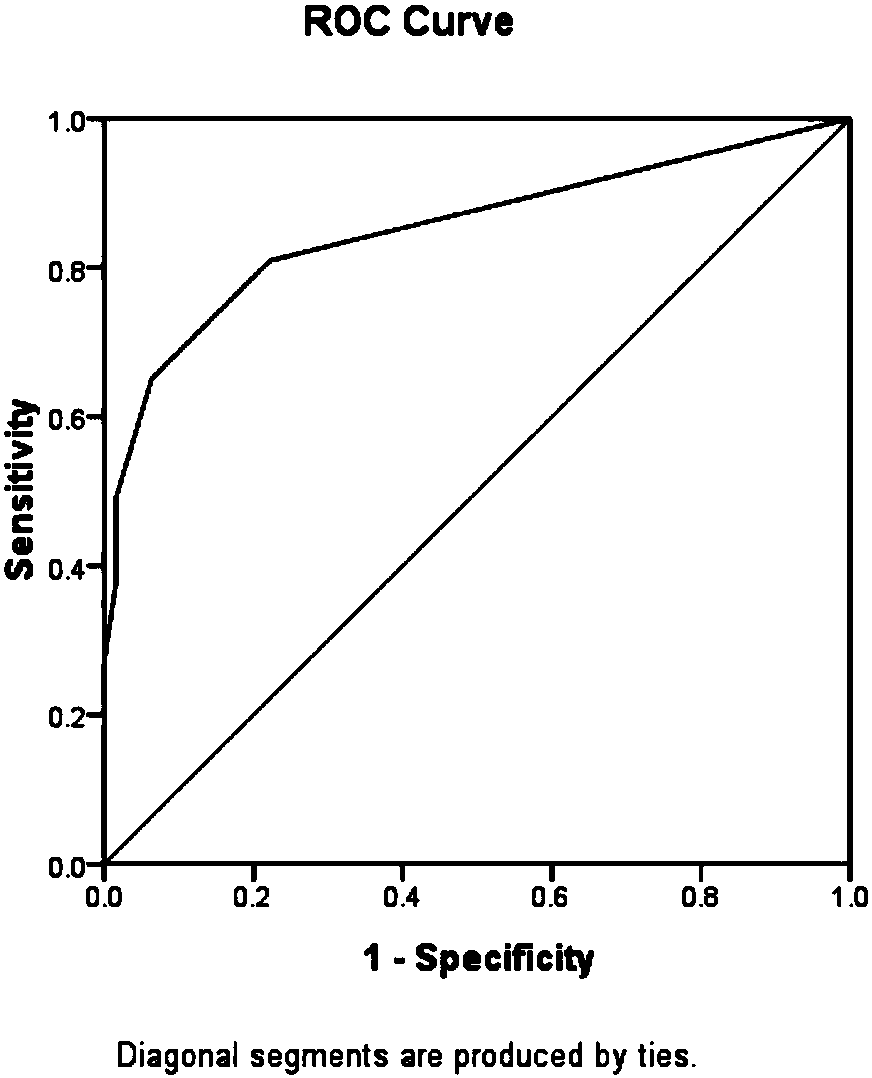
[0186] Taking 4 healthy samples and 8 clinically diagnosed severe oligoasthenospermia samples as the research objects for verification, the serine at the 208th position of the AKAP3 protein and the mass deviation of -113.05347 at the 186th position of the AKAP4 protein were detected respectively in the above samples. Asparagine shifted, asparagine with mass shift of -114.04278 at position 186 of AKAP4 protein, glutamine with mass shift of -17.02660 at position 617 of AKAP4 protein, lysine with mass shift of +211.09682 at position 733 of AKAP4 protein , lysine with +42.01108 mass shift at position 531 of ATP5A1 protein, lysine with +42.01108 mass shift at position 87 of COX4I1 protein, threonine with +79.96685 mass shift at position 64 of GAPDHS protein and 692 position of KIAA1683 protein The respective detection frequencies of serine with +79.96685 mass shift occurred, and the samples to be tested were diagnosed according...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com