Endo-xylanase reorganized mutant having improved salt adaptability and its preparation method and application
An endo-xylanase and mutant technology, which can be used in the fields of genetic engineering and protein modification, and can solve the problems of lack of catalytic activity and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1: Construction of mutant library
[0032] ①The genomes of Arthrobacter sp. and Lechevalieria sp. were extracted according to the instructions of the GENE STAR Bacterial Genome Extraction Kit.
[0033] ②According to the Arthrobacter sp. endoxylanase nucleotide sequence JQ863105 (SEQ ID No.3) recorded in GenBank, primers 5' GTGCAGCCGGAGGAAAAACG 3' and 5' GATGAAGGCAGGATCCGGGGT 3' were designed to target Arthrobacter sp. ) genome as a template for PCR amplification to obtain the endoxylanase gene xynAGN16L; in addition, according to the nucleotide sequence JF745868 (SEQ ID No. 4) Design primers 5'GTCTCGGCCCCGCCGGACGT 3' and 5'GGCTCGCTTCGCCAGCGTGG 3', and perform PCR amplification using the Lechevalieria sp. genome as a template to obtain the endoxylanase gene xynAHJ3.
[0034] ③PCR reaction parameters are: denaturation at 94°C for 5 min; then denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec, extension at 72°C for 1 min and 30 sec, and after 30 cycles,...
Embodiment 2
[0040] Example 2: Screening of Mutants
[0041] 1) Take 2 μL of bacterial liquid from the 96-well cell culture plate in which the mutant library is stored, and inoculate it into 200 μL / well liquid LB culture medium (containing 100 μg mL -1 Amp) in a 96-deep-well plate at 37°C with shaking at 200rpm until OD 600 >1.0 (about 20h), add 2mM IPTG and 100μg mL -1 Amp was induced overnight at 20° C. with 160 rpm in 200 μL liquid LB culture solution.
[0042] 2) Add 40 μL / well of PopCulture after induction TM Cell lysate, at 25°C, shake and lyse the cells for 30 minutes.
[0043] 3) Take 50 μL of McIlvaine buffer (pH=7.0) containing 1.0% (w / v) beech xylan and 50 μL of cell lysate, and react in a 96 deep-well plate in a 70° C. incubator for 2 hours. After the reaction, add 150 μL of DNS reagent to terminate the reaction, incubate in a 140°C incubator for more than 20 minutes and cool to room temperature, and use a microplate reader to read the OD 540nm The value of E.coli BL21-Gol...
Embodiment 3
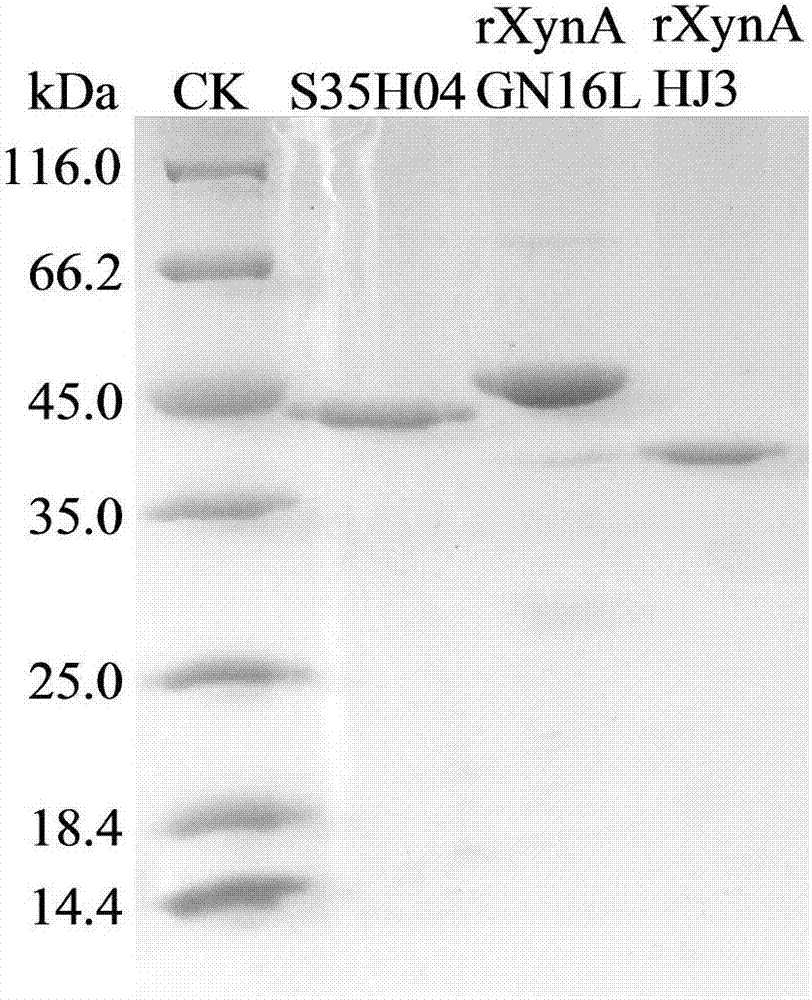
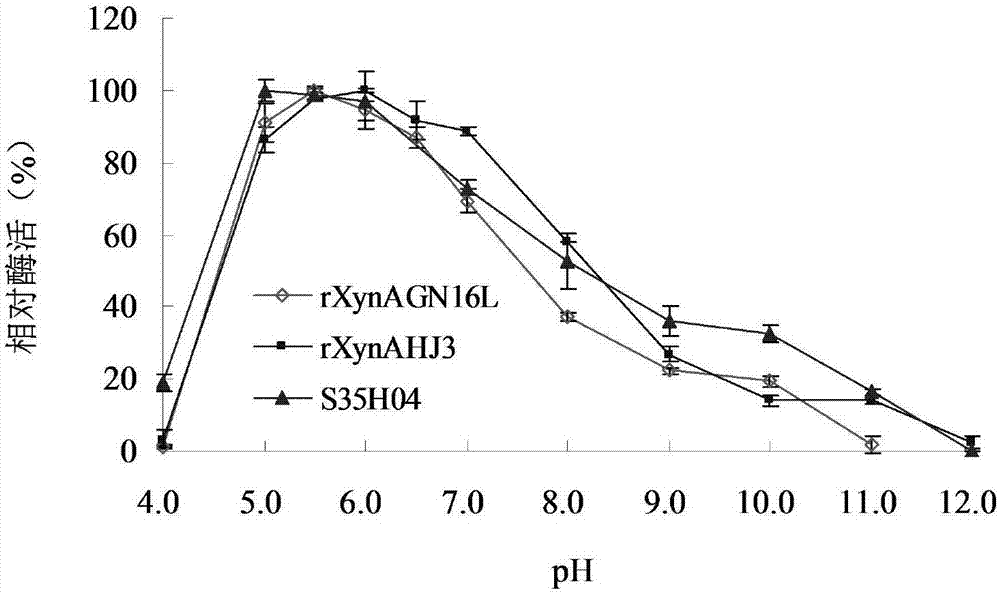
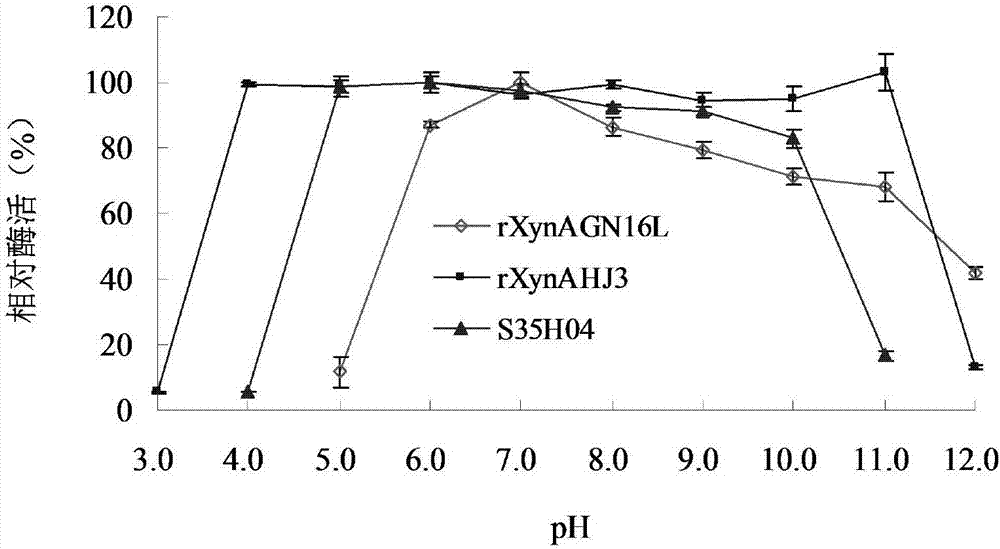
[0047] Embodiment 3: Enzyme preparation of mutant S35H04 and wild enzyme rXynAGN16L and rXynAHJ3
[0048] The recombinant strains containing mutant S35H04, wild enzymes rXynAGN16L and rXynAHJ3 were inoculated in LB (containing 100 μg mL -1 Amp) medium, shake rapidly at 37°C for 16h.
[0049] Then inoculate the activated bacterial solution into fresh LB (containing 100 μg mL -1 Amp) culture medium, rapid shaking culture for about 2–3h (OD 600 After reaching 0.6-1.0), add IPTG at a final concentration of 0.1 mM for induction, and continue shaking culture at 20° C. for about 20 h. Centrifuge at 12000rpm for 5min to collect the bacteria. After suspending the bacteria with an appropriate amount of pH=7.0 Tris-HCl buffer solution, the bacteria were disrupted ultrasonically in a low-temperature water bath. The crude enzyme solution concentrated in the cells above was centrifuged at 13,000rpm for 10min, the supernatant was aspirated and the target protein was affinity-purified wit...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap