A CRISPR/NCas9-mediated site-directed base replacement in plants
A plant and gene technology, applied in the application field of fixed-point base replacement in plants, can solve the problem that agronomic traits cannot be improved quickly
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] Example 1. Application of a CRISPR / nCas9-mediated site-directed base replacement in plants
[0082] 1. Construction of expression vector
[0083] 1. Construction of pCXUN-BE3 vector
[0084] (1) digest the pCXUN-Cas9 vector with restriction endonuclease BamHI to obtain a linearized vector;
[0085] (2) PCR amplification was carried out with BE-F / R as primers and pCMV-BE3 vector as template to obtain a PCR product. The sequences at the 5' and 3' ends of the PCR product were completely consistent with the sequences at both ends of the linearized vector, respectively. ;
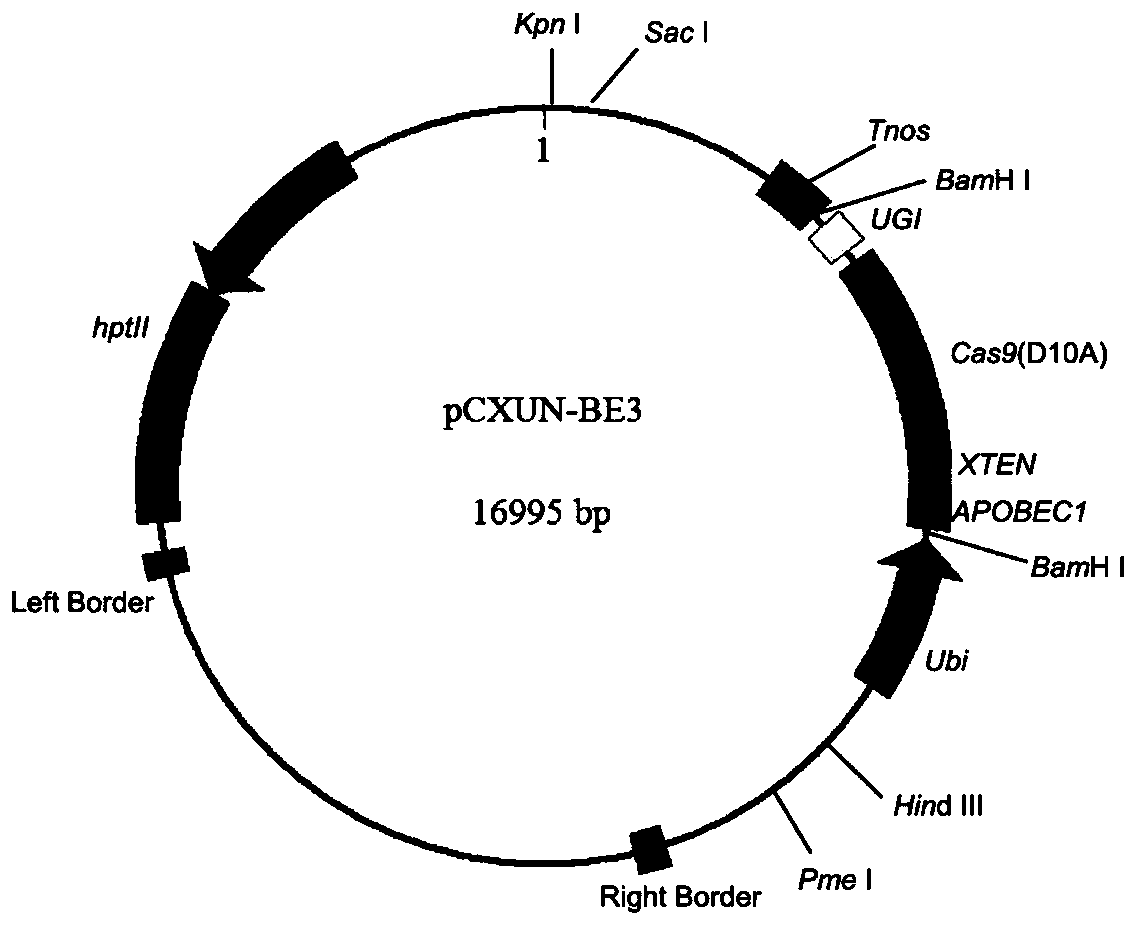
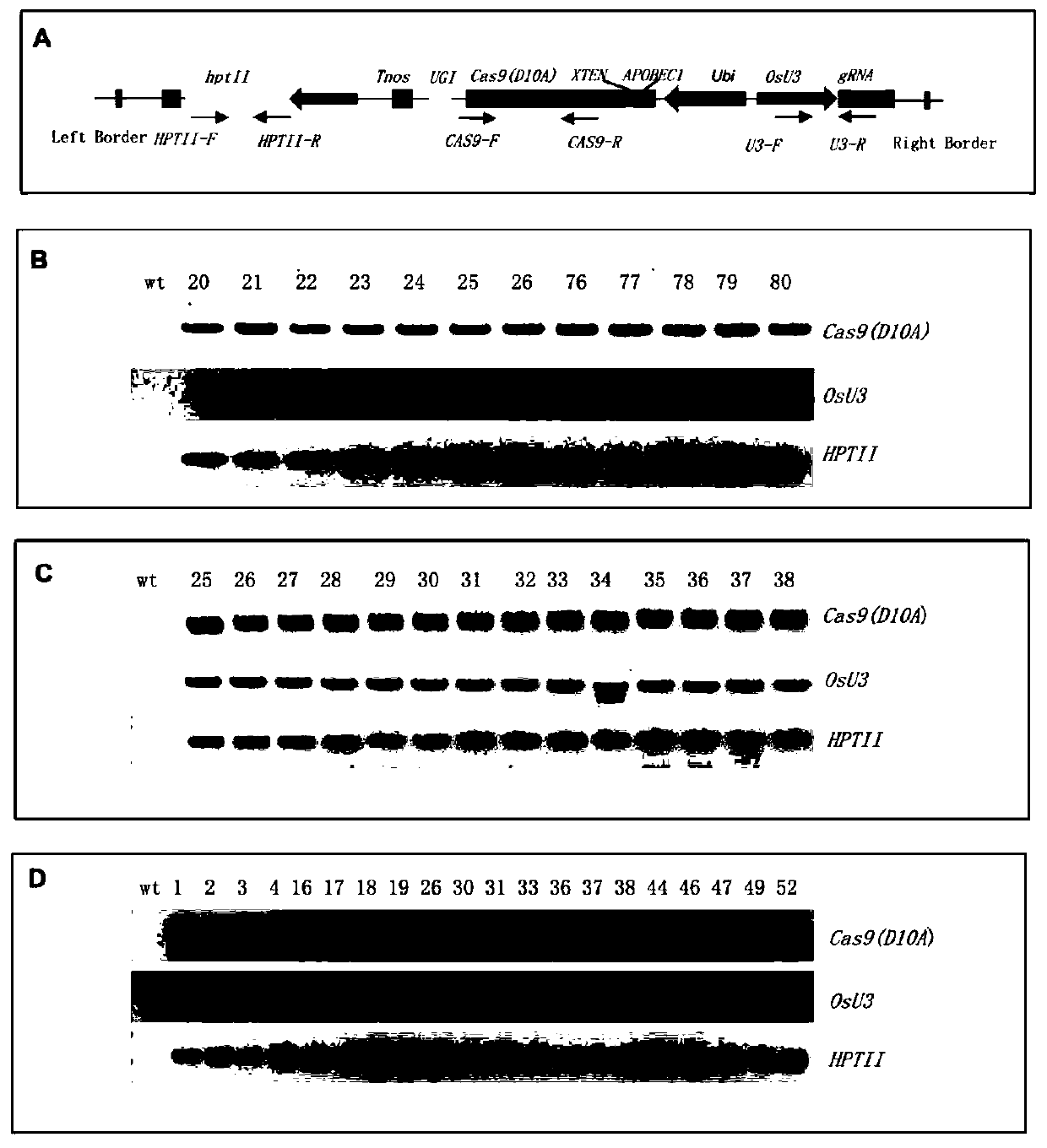
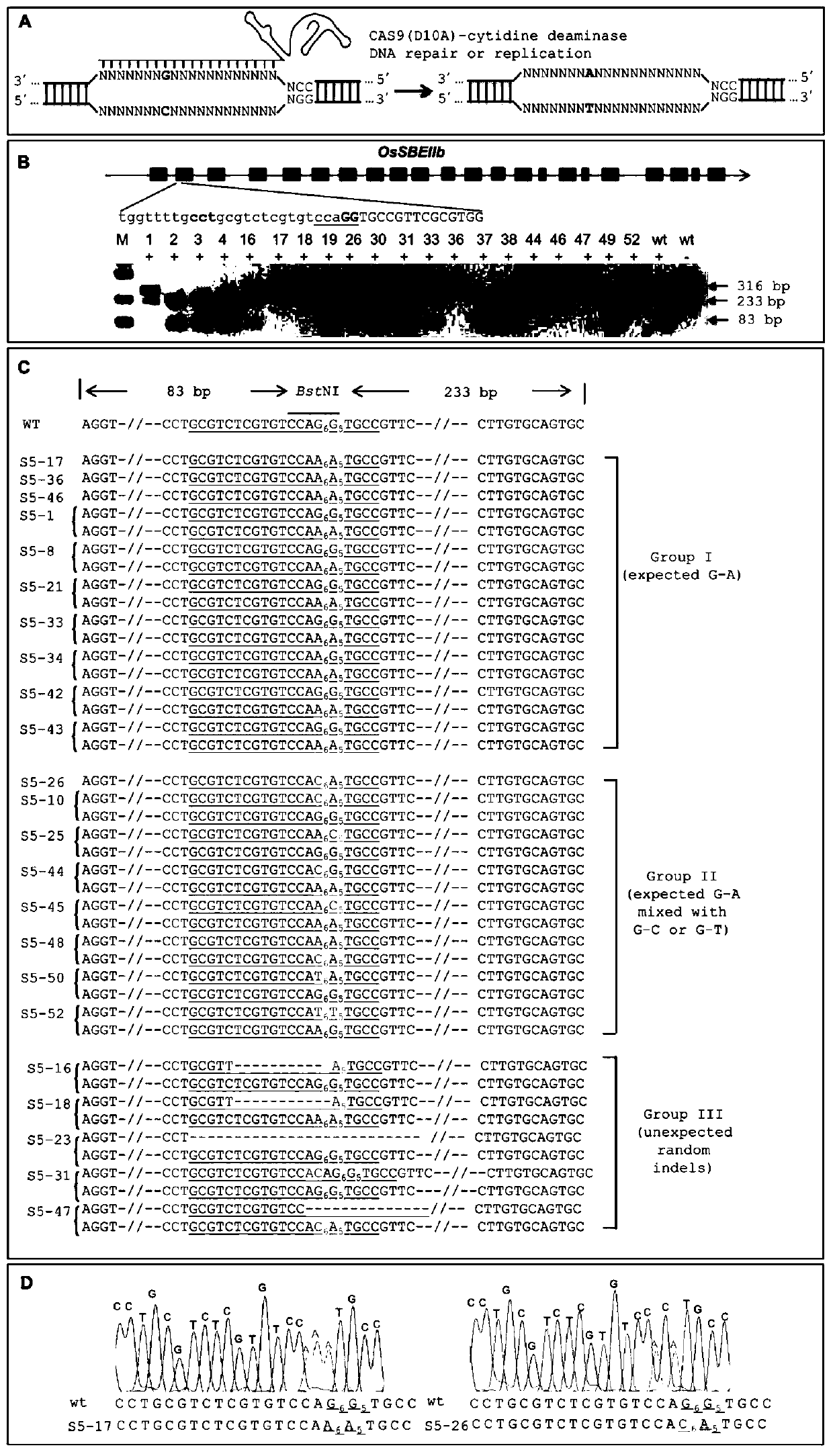
[0086] (3) The linearized vector obtained in step (1) and the PCR product obtained in step (2) were connected by homologous recombination using the pEASY-Uni Seamless Cloning and Assembly Kit of Quanshijin Company to obtain the vector pCXUN-BE3 ( figure 1 ), as can be seen from the figure: the pCXUN-BE3 vector includes an expression cassette A, which in turn includes the maize Ubiquitin promoter, the c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com