A kind of circular RNAcircbcbm1 and its non-diagnostic fluorescence quantitative detection method
A fluorescent quantitative detection, circular technology, used in DNA/RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
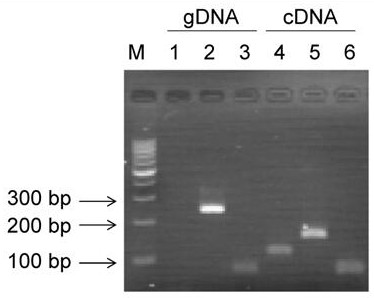
[0056] Circular properties of circular RNA circBCBM1:
[0057] ①Primer design and synthesis: design reverse specific primers for circBCBM1 spanning the splice site, the upstream primer is ctgggccact gatgactcct, the downstream primer is tcctggacac ggtagctcca; the linear transcript primer corresponding to circBCBM1 is designed, the upstream primer is tggagctacc gtgtccagga tg, and the downstream primer is gcggctcgatgaagctgtgg; design primers for internal reference gene GAPDH, the upstream primer is tgagaacggg aagcttgtca, and the downstream primer is atcgccccac ttgattttgg;
[0058] ② DNA and RNA extraction and reverse transcription: culture 231-BR cells, extract the whole genomic DNA (gDNA) of the cells, use TRIzol to extract the total RNA of the cells, and reverse transcribe the total RNA into cDNA;
[0059] Wherein the extraction of RNA can be carried out according to the following steps:
[0060] ⑴Add TRIzol to the cell culture plate to lyse the cells, every 10cm 2 Add 1ml to...
Embodiment 2
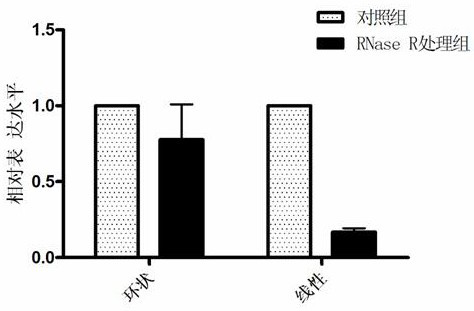
[0076] Stability of circular RNA circBCBM1:
[0077] ① RNA extraction, processing and reverse transcription: 231-BR cells were cultured, and total RNA was extracted using TRIzol. Add 3U RNase R to 2 μg RNA in the RNase R treatment group, add an equal volume of DEPC water to the control group, and treat at 37°C for 30 minutes; purify the RNA and perform reverse transcription;
[0078] Wherein the extraction of RNA and the process of reverse transcription are the same as in Example 1;
[0079] ②Real-time fluorescent quantitative PCR amplification: the reaction was carried out on Applied Biosystems 7500Real-Time PCR instrument;
[0080] First, prepare the PCR reaction solution according to the following components (the reaction solution is prepared on ice)
[0081] Reagent Usage amount Final concentration TB Green Premix Ex Taq II (Tli RNaseH Plus) (2×) 10μl 1× PCR Forward Primer (10μM) 0.8μl 0.4μM PCR Reverse Primer (10μM) 0.8μl 0.4μM ...
Embodiment 3
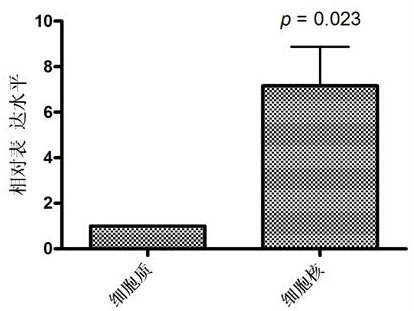
[0086] Subcellular localization of circular RNA circBCBM1:
[0087] The subcellular localization of circular RNA circBCBM1 was detected by real-time fluorescent quantitative PCR. The cytoplasm and nucleus were separated using the NE-PER kit, and the total RNA in the cytoplasm and nucleus were extracted using TRIzol. After reverse transcription, the expression levels of circBCBM1 in the cytoplasm and nucleus were detected by real-time fluorescent quantitative PCR. The RNA extraction and reverse transcription processes were the same. Example 1, the reaction system and conditions of real-time fluorescent quantitative PCR refer to Example 2, GAPDH is used as an internal reference gene.
[0088] The result is as image 3 As shown, the relative expression level of circBCBM1 in the nucleus is 7.16 times higher than that in the cytoplasm, indicating that the subcellular localization of circBCBM1 is mainly in the nucleus.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com