Method and reagent for quantifying cholesterol in triglyceride-rich lipoprotein
A technology for triglycerides and lipoproteins, used in biochemical equipment and methods, microbial determination/inspection, measurement devices, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] The reagent composition A for carrying out the step (1) (the reagent composition for carrying out the step (1) is also referred to as "reagent composition A" hereinafter in Example 2) was prepared as follows, and the reagent composition A for carrying out the step (2) was prepared as follows. (The reagent composition used to carry out step (2) is also referred to as "reagent composition B" after Example 2).
[0071] Reagent composition A
[0072] PIPES buffer, pH6.8 50mmol / L
[0073] Various cholesterol esterases (refer to Table 1) 3U / mL
[0074] Cholesterol oxidase 3U / mL
[0075] Catalase 1200U / mL
[0076]TOOS 2.0mmol / L
[0077] Polyoxyethylene stilbene phenyl ether [HLB:12.8] 0.25%(w / v)
[0078] Reagent composition B
[0079] PIPES buffer, pH6.8 50mmol / L
[0080] 4-Aminoantipyrine 4.0mmol / L
[0081] Peroxidase 20 units / mL
[0082] Sodium azide 0.05%(w / v)
[0083] Polyoxyethylene alkyl ether 0.5%(w / v)
[0084] 150 μL of reagent composition A was added to 3 μ...
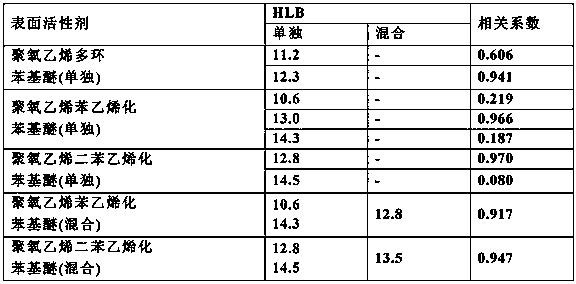
Embodiment 2
[0090] Reagent Composition A and Reagent Composition B were formulated as described below.
[0091] Reagent composition A
[0092] PIPES buffer, pH6.8 50mmol / L
[0093] Cholesterol esterase [62kDa] 3U / mL
[0094] Cholesterol oxidase 3U / mL
[0095] Catalase 1200U / mL
[0096] TOOS 2.0mmol / L
[0097] Various surfactants (refer to Table 2)※ 0.25%(w / v)
[0098] ※In the case of combining 2 or more types, the total is 0.25% (w / v)
[0099] Reagent composition B
[0100] PIPES buffer, pH6.8 50mmol / L
[0101] 4-Aminoantipyrine 4.0mmol / L
[0102] Peroxidase 20 units / mL
[0103] Sodium azide 0.05%(w / v)
[0104] Polyoxyethylene alkyl ether 0.5%(w / v)
[0105] 150 μL of reagent composition A was added to 3 μL of serum sample, and after reacting at 37° C. for 5 minutes, 50 μL of reagent composition B was added and allowed to react for 5 minutes, and the absorbance at the main wavelength of 600 nm and the sub-wavelength of 700 nm was measured. Table 2 shows the correlation coeffici...
Embodiment 3
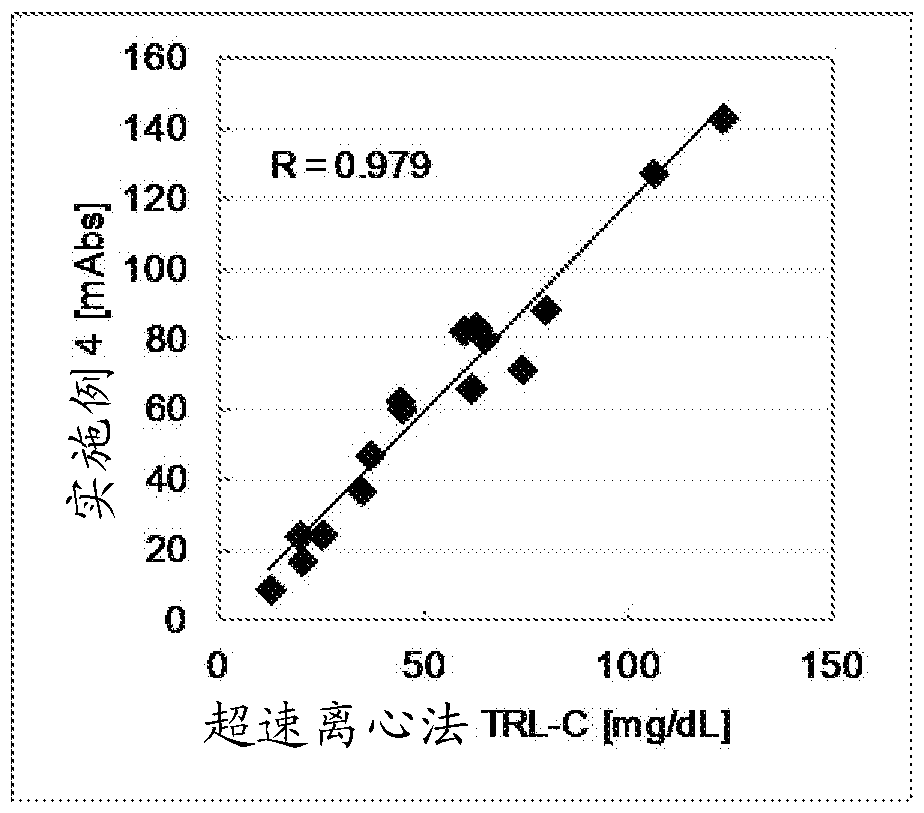
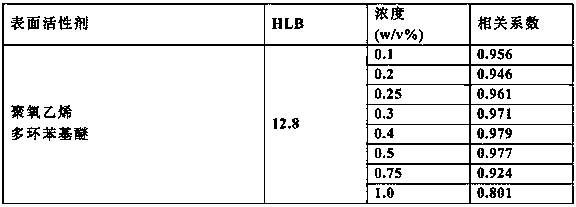
[0110] Reagent Composition A and Reagent Composition B were formulated as described below.
[0111] Reagent composition A
[0112] PIPES buffer, pH6.8 50mmol / L
[0113] Cholesterol esterase [62kDa] 3U / mL
[0114] Cholesterol oxidase 3U / mL
[0115] Catalase 1200U / mL
[0116] TOOS 2.0mmol / L
[0117] Polyoxyethylene polycyclic phenyl ether [HLB: 12.8] Various concentrations (refer to Table 3)
[0118] Reagent composition B
[0119] PIPES buffer, pH6.8 50mmol / L
[0120] 4-Aminoantipyrine 4.0mmol / L
[0121] Peroxidase 20 units / mL
[0122] Sodium azide 0.05w / v%
[0123] Polyoxyethylene alkyl ether 0.5w / v%
[0124] 150 μL of reagent composition A was added to 3 μL of serum sample, and after reacting at 37° C. for 5 minutes, 50 μL of reagent composition B was added and allowed to react for 5 minutes, and the absorbance at the main wavelength of 600 nm and the sub-wavelength of 700 nm was measured. Table 3 shows the correlation coefficient obtained by comparing with the TRL-...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| density | aaaaa | aaaaa |
| density | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com