Internal reference gene hsa_circ_0000471 of human tissue/cell circular RNA and its application
A technology of human tissue and internal reference genes, applied in the field of molecular biology, can solve problems such as the inability to rule out the interference of linear shear isomers and correct the expression of target genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] 1. Screen candidate internal reference genes for circular RNA
[0018] Log in to the Gene Expression Omnibus (GEO) website (https: / / www.ncbi.nlm.nih.gov / geo / ) to download the dataset numbered GSE77661 (GSE77661_circ_srpbm_union_samples_loci.txt.gz), which includes 6 healthy human specimens (brain, colon, heart, liver, lung, and stomach) and 7 pairs of cancerous and paracancerous human tissue specimens (bladder, breast, colorectal, gastric, liver, clear cell, and prostate) circRNA-sequencing result.
[0019] The data processing method is as follows:
[0020] (1) Only the circular RNAs expressed in all samples were retained, and a total of 50 circular RNAs were obtained.
[0021] (2) Analyze the stability of the 50 circular RNAs obtained in tissue samples using geNorm, NormFinder and BestKeeper respectively, take the 10 circular RNAs with the best stability in each method, and then combine the obtained 3 data Set the intersection and get 6 circular RNAs.
[0022] 2. F...
Embodiment 2
[0038] Evaluation of specificity and amplification efficiency of hsa_circ_0000471
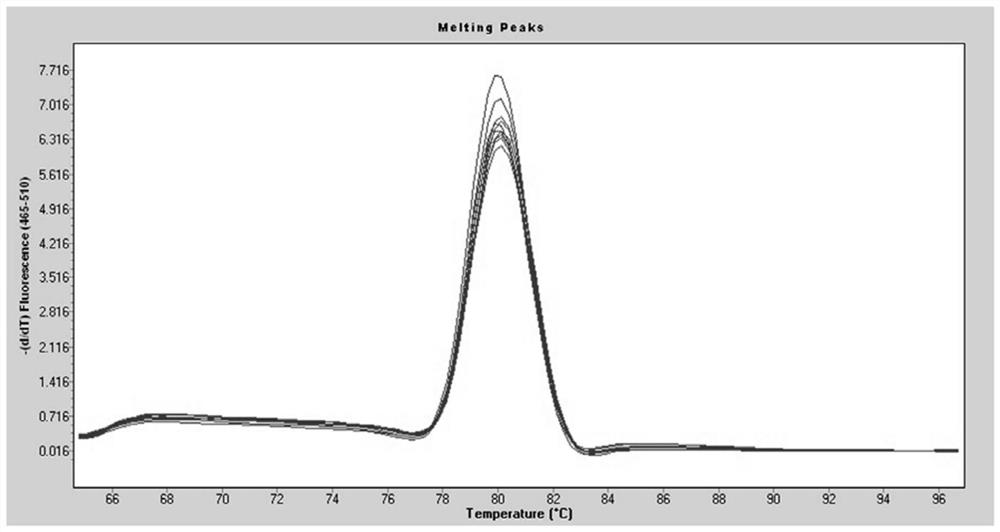
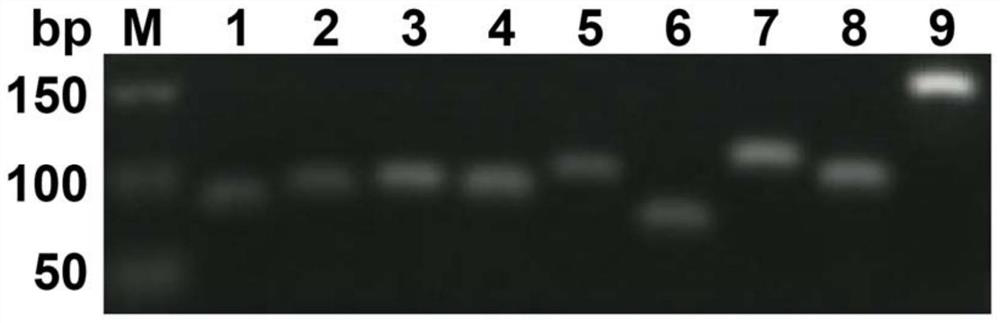
[0039] The results of RT-qPCR showed that the melting curve of hsa_circ_0000471 was a single peak ( Figure 1a shown), indicating that there is no non-specific amplification. In order to further confirm that there is no amplification of non-specific bands, 2% agarose gel electrophoresis was used to amplify the product, and the results were as follows: Figure 1b As shown, no miscellaneous bands were seen.
[0040] In order to obtain the amplification efficiency of hsa_circ_0000471, the template was first diluted, and then amplified on the machine. After the amplification result was obtained, the result was fitted with a straight line to obtain the slope (slope), and then the amplification efficiency was calculated using the following formula:
[0041] E(%)=(2 -1 / slope -1)×100
[0042] Finally, the amplification efficiency of hsa_circ_0000471 was found to be 99.93%.
PUM
| Property | Measurement | Unit |
|---|---|---|
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com