Integration sites in CHO cells
A technology of cells and Chinese hamsters, applied in the field of identification of genomic integration sites, can solve the problem of difficult prediction of the possibility of genomic loci
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0151] CHO producer cell clones are usually obtained by random integration of heterologous polynucleotides into the host cell genome of CHO cells, ie by random integration (RI). The position effect results in a highly heterogeneous cell population consisting mainly of low producer cells with only a small subset of high producer cells. Additionally, high producer cells tend to be outnumbered by low producer cells. To assess the potential of the Chinese hamster S100A gene cluster as a reliable, high-level production site (i.e., a "hotspot") for heterologous protein production, DNA engineered to SEQ ID NO: 11 (ZFN 13) was used as described above. Sequence-specific pairs of zinc finger nucleases that stably integrate polynucleotides encoding IgG antibodies into the genomes of CHO-DG44 and CHOZN GS cells.
[0152] After confirmation of ZFN activity and preparation of donor plasmids, cells were co-transfected with a non-linear plasmid containing an expression cassette encoding an I...
Embodiment 2
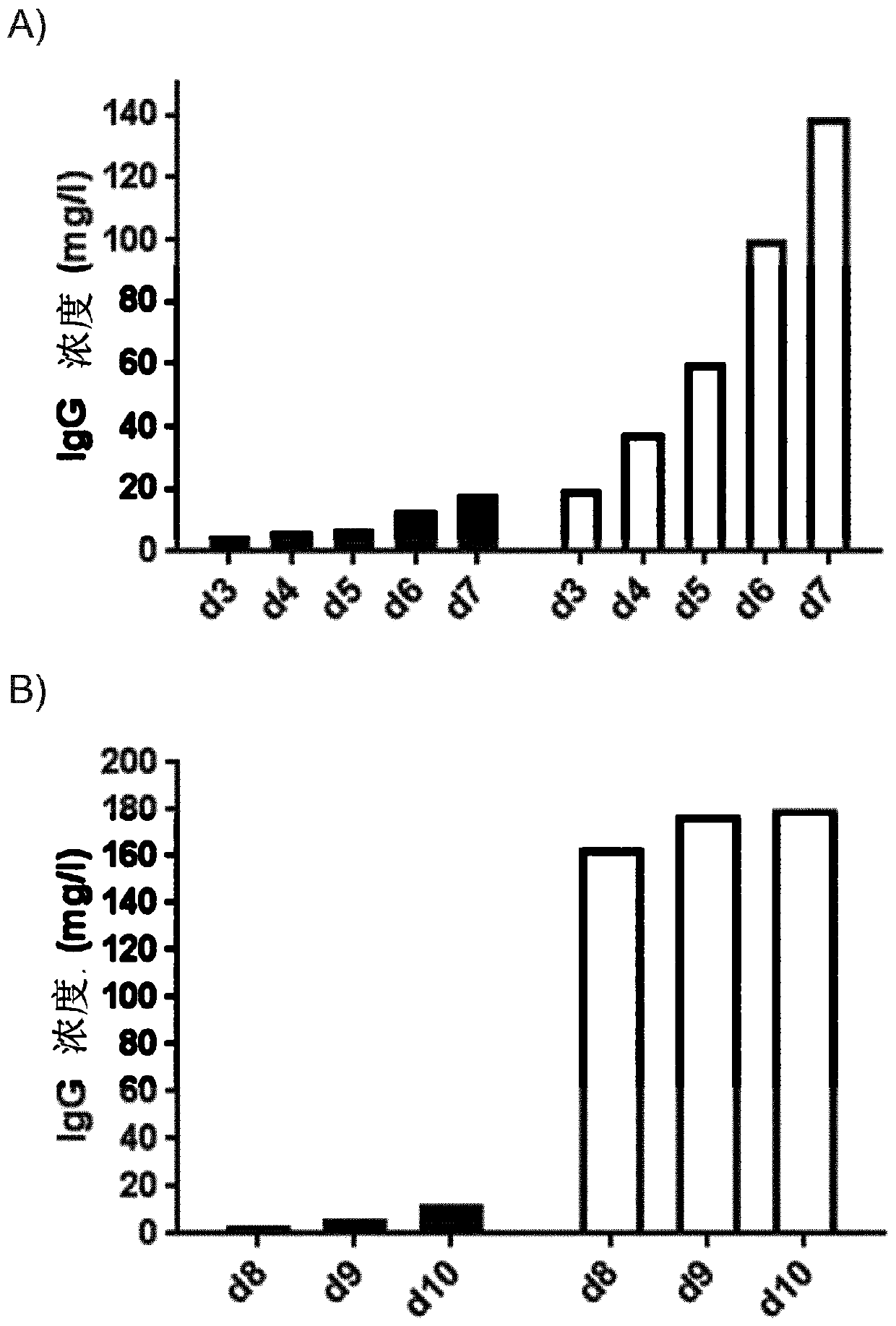
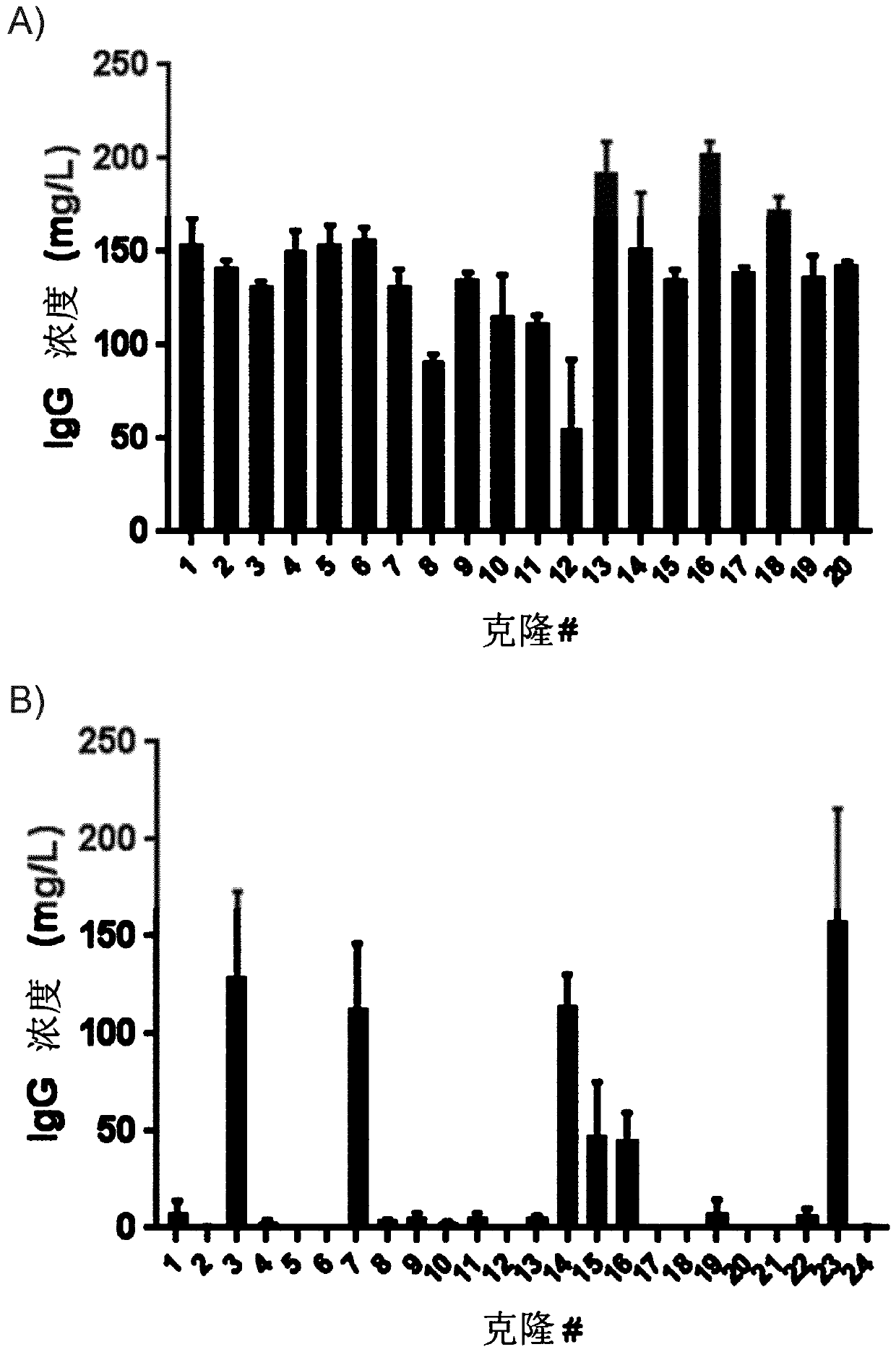
[0157] Random integration results in cell repertoires that are highly heterogeneous in expression of heterologous proteins. To assess whether targeted integration within the Chinese hamster S100A gene cluster resulted in more uniform expression levels and thus greater predictability in terms of productivity, individual clones were selected from the TI and RI cell banks of Example 1.
[0158] Single cell clones (SCC) were obtained using targeted integration and random integration pools from CHOZN GS cells from Example 1. A single cloning process was accomplished by limiting the dilution of enriched TI and RI pools using conditioned medium. Prepare conditioned media by culturing cells at 0.3e6 cells / ml for 48 h in TPP tubes. Cells were pelleted and conditioned medium was sterile filtered. Inoculation was performed in an 80:20 mixture of cloning medium (SAFC fusion platform) and conditioned medium using the following procedure. Step 1: Place serial dilutions in a 96-well plate...
Embodiment 3
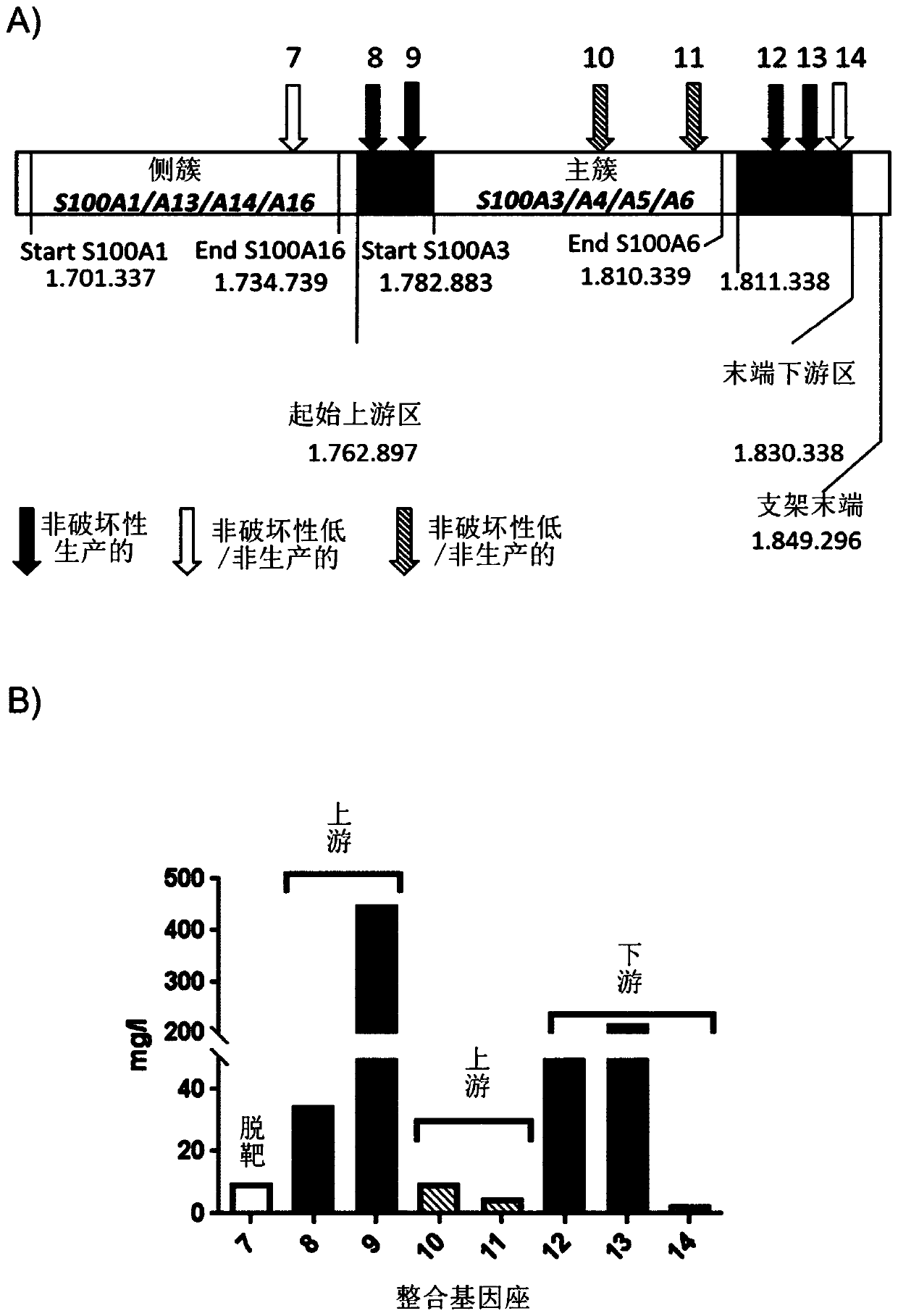
[0162] To validate the hotspot loci in the S100A gene cluster, a number of additional TI zinc finger nucleases were designed and generated as shown in Table 3 to create a production library as described in Example 1. image 3 A shows the positions of individual ZFNs and hotspot loci in the S100A gene cluster with NCBI standard sequence: NW_003613854.1. Shown are the integration sites of ZNF 7 to 14, categorized as "non-destructive and productive", "non-destructive and low / non-productive" and "destructive and low / non-productive" sites.
[0163] Data were generated using CHO-ZN GS cells as described in Example 1. Eight different genomic loci were tested to assess whether a region favors heterologous gene product production relative to the S100A3 / A4 / A5 / A6 major gene cluster. It was further tested whether integration into the S100A3 / A4 / A5 / A6 major gene cluster would result in reduced productivity ( image 3 B).
[0164] table 3:
[0165] zinc finger nuclease target...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com