Application of a circRNA marker for the diagnosis of colorectal cancer
A colorectal cancer and marker technology, applied in the application field of circRNA markers, can solve the problems of missed diagnosis and misdiagnosis, low sensitivity and specificity, and too expensive in colorectal cancer detection, achieving strong sensitivity, stable results, Strong effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
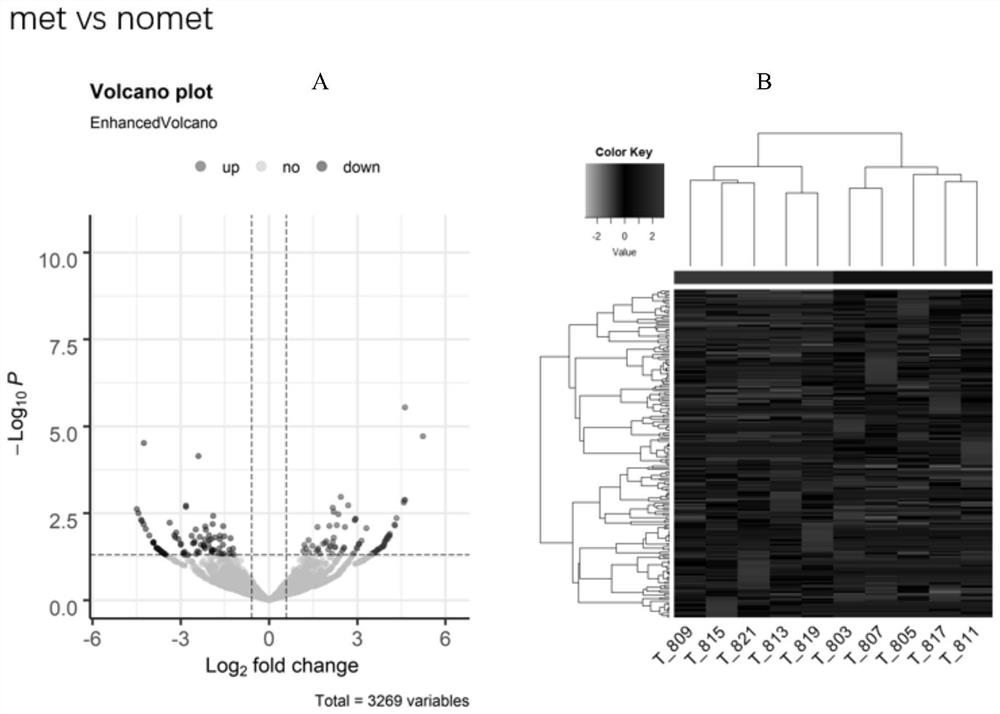
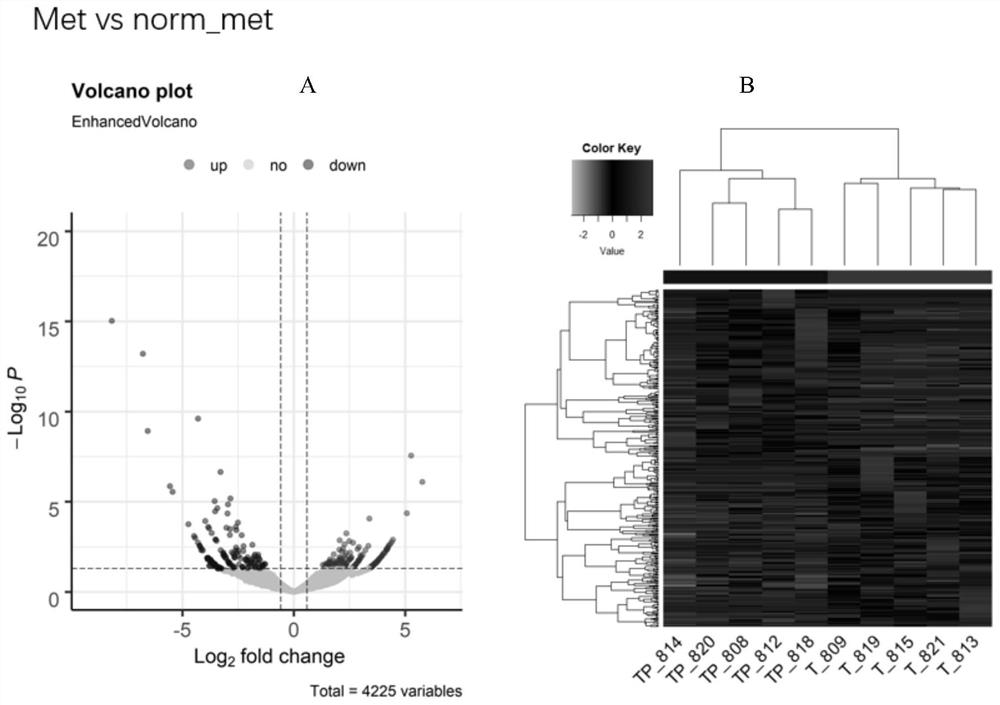
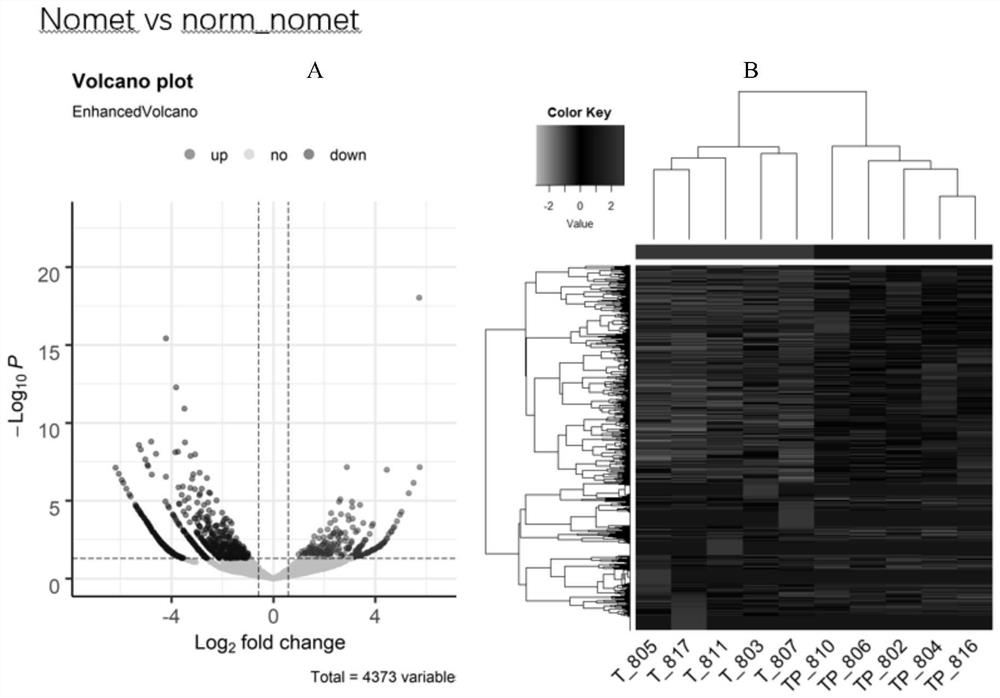
[0038] Ten pairs of colorectal cancer tissues (carcinoma and paracancerous) were divided into two groups: metastatic group (5 pairs) and non-metastatic group (5 pairs). The metastatic group is further divided into a metastatic cancer group (denoted by Met) and a metastatic paracancer group (denoted by norm_met), and a non-metastatic group is divided into a non-metastatic cancer group (denoted by nomet) and a non-metastatic paracancerous group (denoted by norm_nomet). Through high-throughput sequencing technology, all differentially expressed circRNAs between the two groups were screened out. The result is as Figure 1~3 As shown, there are 187 differentially expressed circRNAs between the Met group and the nomet group, 87 of which are up-regulated and 100 are down-regulated; there are 350 differentially expressed circRNAs between the Met group and the norm_met group, of which 176 Up-regulated and 174 down-regulated; there were 1036 differentially expressed circRNAs between th...
Embodiment 2
[0042] All differentially expressed circRNAs screened were analyzed by circRNA high-throughput sequencing technology, and the intersection between each group was taken. 结果发现同时在肿瘤组vs癌旁组和Met vs norm_met组两组间差异表达的circRNA有12种(hsa_circ_0057123,hsa_circ_0000707,hsa_circ_0005704,hsa_circ_0004456,hsa_circ_0005379,hsa_circ_0130312,hsa_circ_0006672,hsa_circ_0001917,hsa_circ_0087641,hsa_circ_0003855,hsa_circ_0018992和hsa_circ_0008230) ; At the same time, there are two circRNAs (hsa_circ_0003761, hsa_circ_0133953) that are differentially expressed between the Met vsnomet group and the Met vs norm_met group, hsa_circ_0133953 was discarded because its sequence was too long, and finally hsa_circ_000376 was selected as the object of further research. Subsequently, Sanger sequencing was used to verify that hsa_circ_0003761 was derived from the four exons 9, 10, 11, and 12 of the MSH3 gene, with a length of 423 bp circular structure ( Figure 4 ). The sequence of hsa_circ_0003761 is shown in SEQ ...
Embodiment 3
[0053] cDNA was obtained by extracting RNA with circRNA primers and performing reverse transcription. Agarose gel electrophoresis obtained by PCR amplification using cDNA as a template showed that only one band was visible after electrophoresis analysis. Lane1 was the DS2000 marker, and the gene fragment represented by Lane2 was hsa_circ_0003761 The PCR amplification product of the gene fragment is between 100bp and 250bp in size, which is consistent with the designed primer sequence, and the results are as follows Figure 5 As shown in A. In order to verify the identity of the sequence of the qRT-PCR product with the target circRNA, we performed reverse sequencing verification on the product sequence. The sequencing results were compared with the Circbase database hsa_circ_0003761 Circularization site sequence verification, the results are as follows Figure 5 B shows that the amplified product is consistent with the target hsa_circ_0003761 sequence, indicating that the ampl...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com