Patents
Literature
1283 results about "Rectal carcinoma" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Compositions and methods for the treatment of cancer
InactiveUS20020128228A1Reducing and avoiding adverse effectImprove toleranceBiocideAnimal repellantsIntestinal structureCancer prevention
This invention relates to compositions comprising temozolomide and thalidomide which can be used in the treatment or prevention of cancer, in particular malignant melanoma, cancer of the skin, subcutaneous tissue, lymph nodes, brain, lung, liver, bone, intestine, colon, heart, pancreas, adrenals, kidney, prostate, breast, colorectal, or a combination thereof. A particular composition comprises temozolomide, or a pharmaceutically acceptable salt, solvate, or clathrate thereof, and thalidomide, or a pharmaceutically acceptable salt, solvate, or clathrate thereof. The invention also relates to methods of treating or preventing cancer, in particular malignant melanoma, cancer of the skin, subcutaneous tissue, lymph nodes, brain, lung, liver, bone, intestine, colon, heart, pancreas, adrenals, kidney, prostate, breast, colorectal, or a combination thereof, which comprise the administration of temozolomide and thalidomide and another anti-cancer drug to a patient in need of such treatment or prevention. The invention further relates to methods of reducing or avoiding adverse side effects associated with the administration of cancer chemotherapy or radiation therapy which comprise the administration of temozolomide and thalidomide to a patient in need of such reduction or avoidance.
Owner:MEMORIAL SLOAN KETTERING CANCER CENT
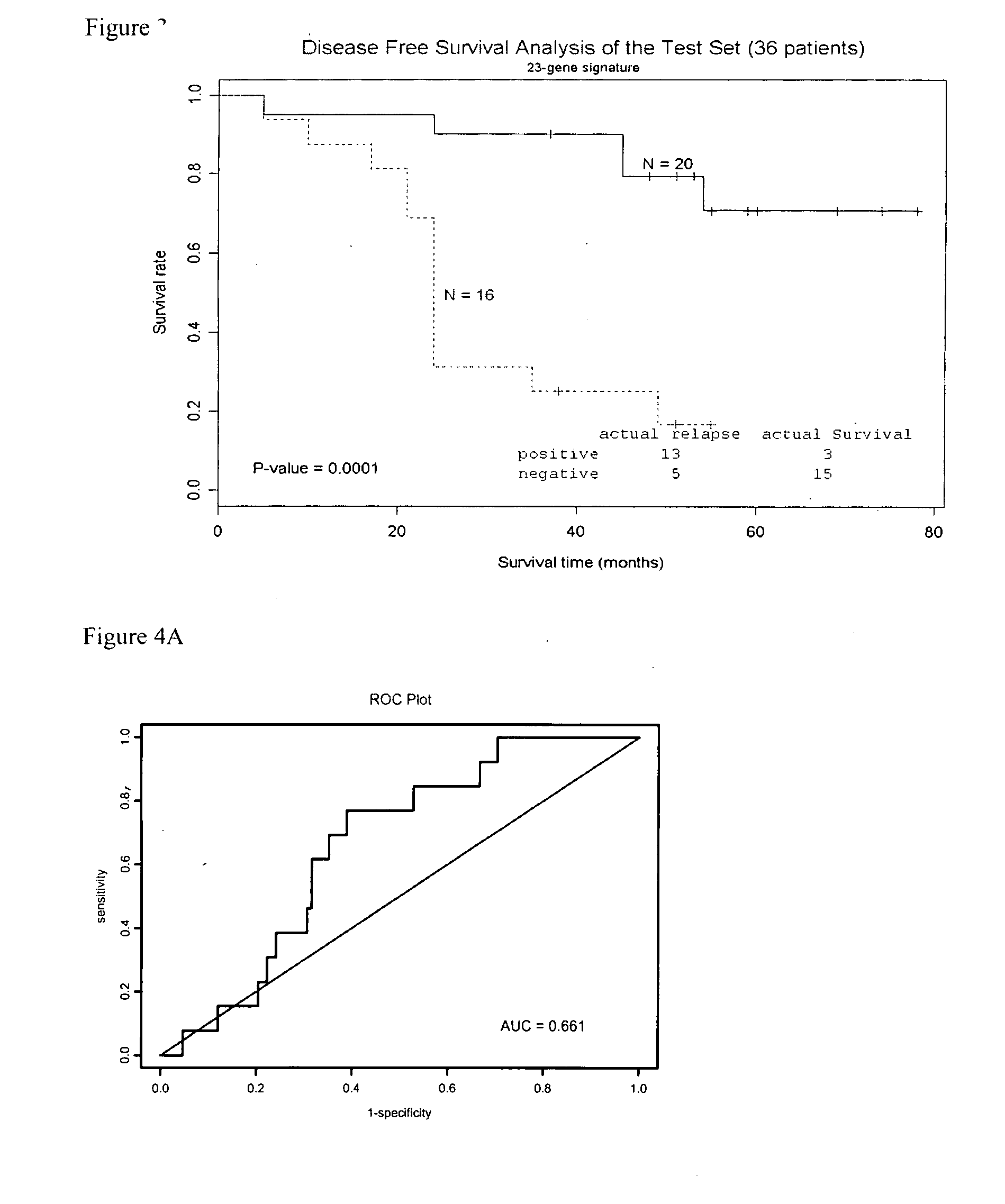
Molecular assay to predict recurrence of Duke's B colon cancer
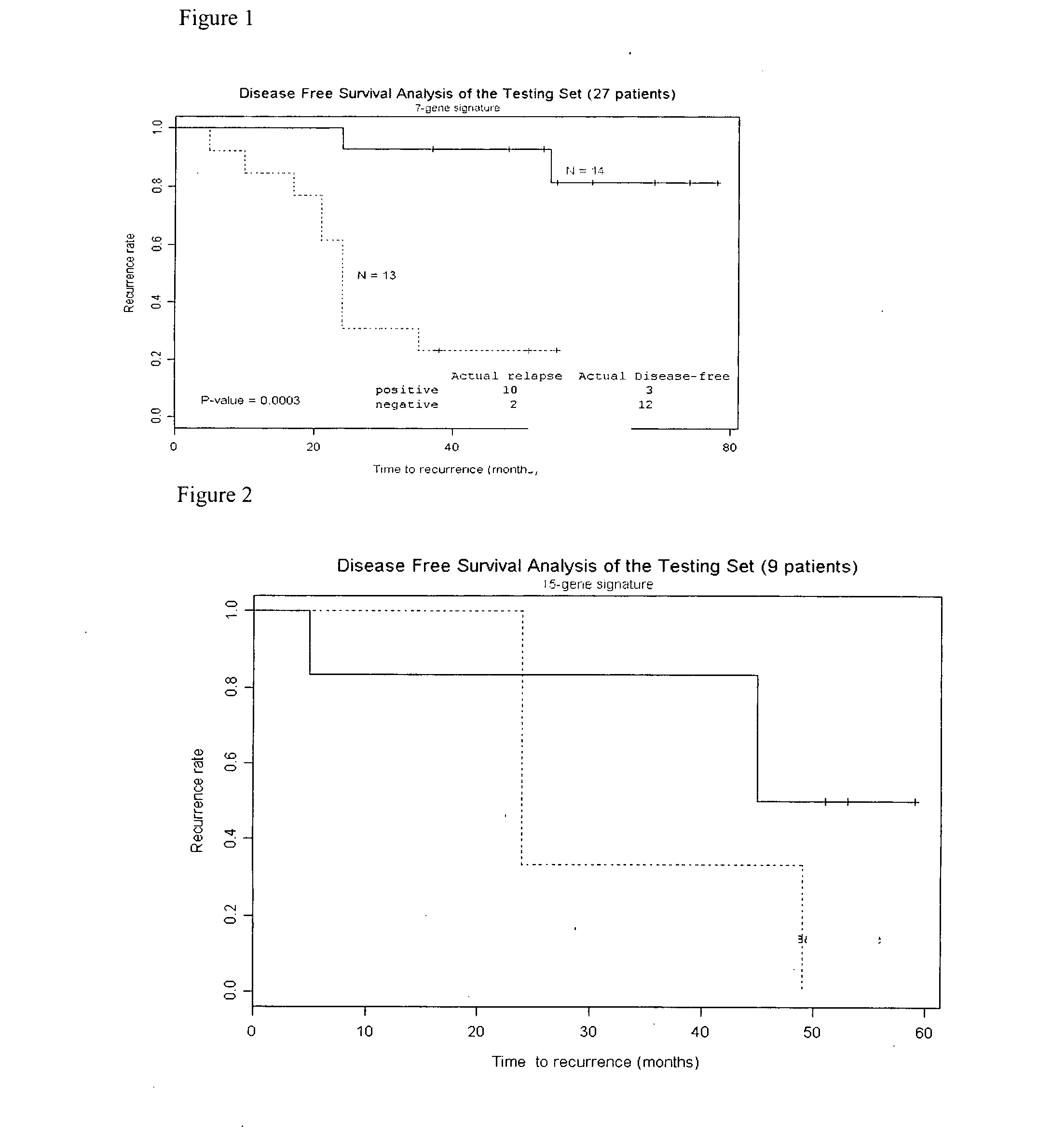
A method of providing a prognosis of colorectal cancer is conducted by analyzing the expression of a group of genes. Gene expression profiles in a variety of medium such as microarrays are included as are kits that contain them.
Owner:VERIDEX LCC
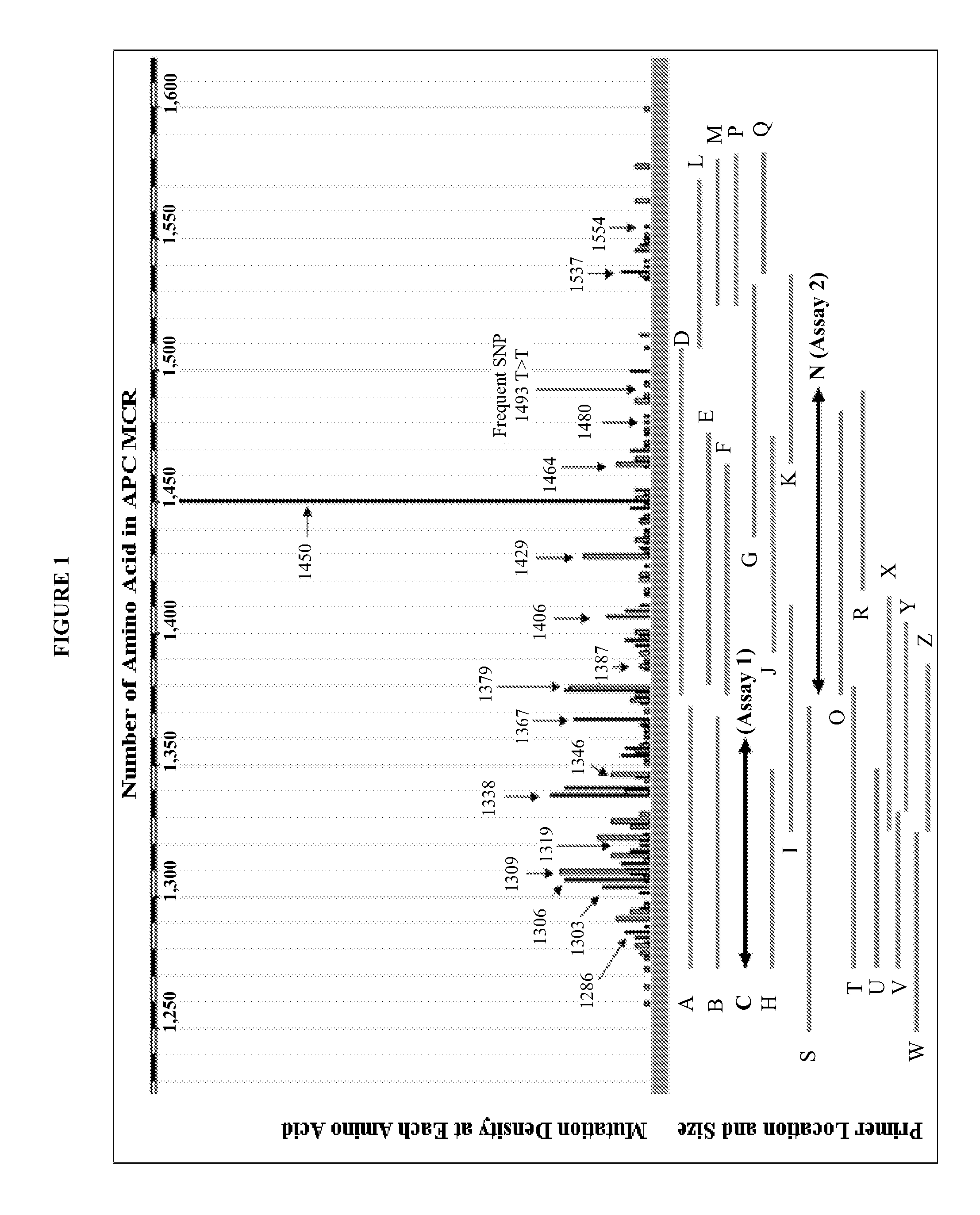
Mutations in human MLH1 and human MSH2 genes useful in diagnosing colorectal cancer
Variant human MLH1 and MSH2 genes are provided. Methods of using these variant genes to diagnose hereditary non-polyposis colorectal cancer (HNPCC) and / or determine a patient's susceptibility to developing HNPCC are also provided. Methods and compositions for identifying new variant MLH1 of MSH2 genes are also provided. In addition, experimental models for hereditary non-polyposis colorectal cancer comprising these variant genes are provided.
Owner:DIADEXUS
Novel peptides and combination of peptides and scaffolds thereof for use in immunotherapy against colorectal carcinoma (CRC) and other cancers
ActiveUS20160346371A1Reduce releaseImprove discriminationImmunoglobulin superfamilyTumor rejection antigen precursorsEpitopeMajor histocompatibility
Owner:IMMATICS BIOTECHNOLOGIES GMBH
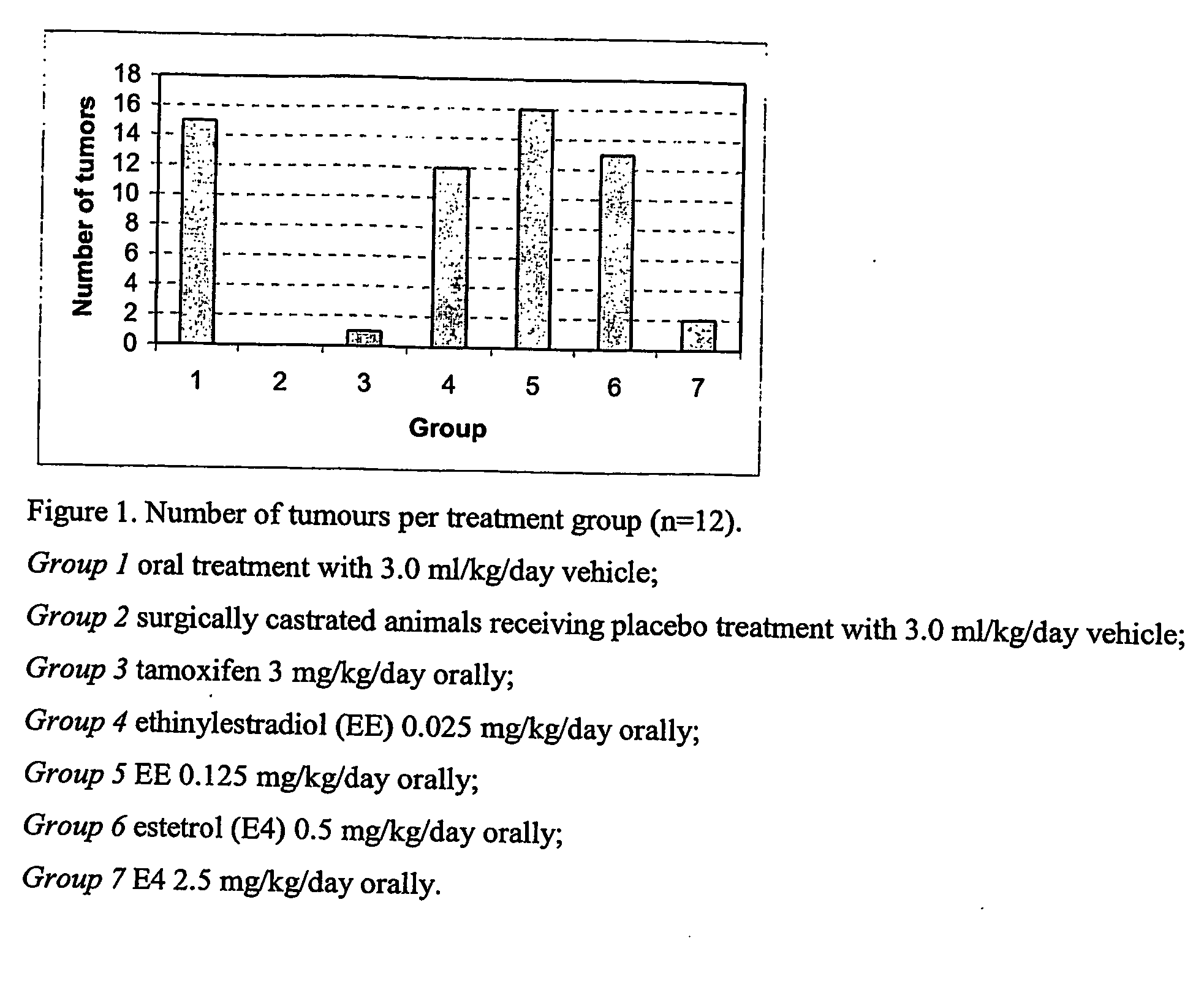
Pharmaceutical compositions comprising estetrol derivatives for use in cancer therapy
ActiveUS20060247221A1High affinityIncreased riskOrganic active ingredientsAntineoplastic agentsGynecologyPresent method
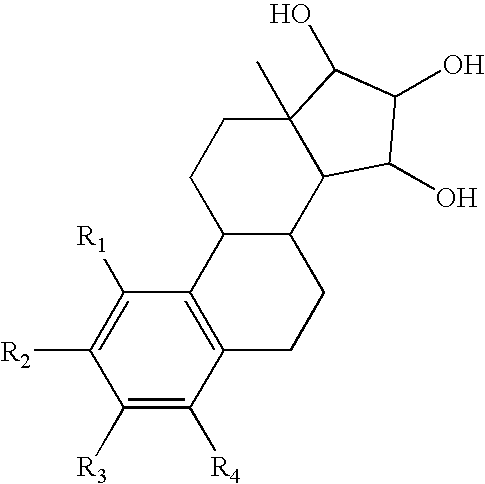
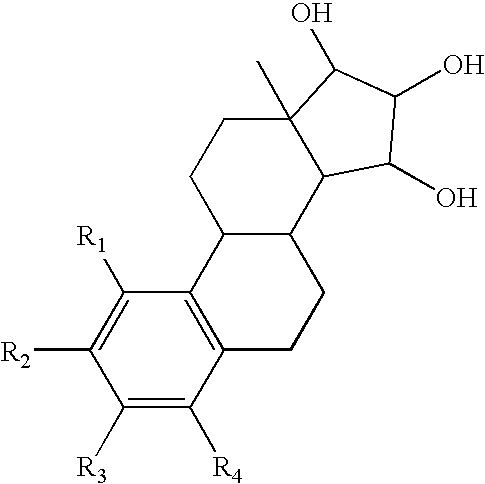
The present invention relates to a method of treating or preventing estrogen-suppressed tumours in a mammal, said method comprising the administration of a therapeutically effective amount of an estrogenic component to said mammal, wherein the estrogenic component is selected from the group consisting of: substances represented by the following formula (I) in which formula R1, R2, R3, R4, independently are a hydrogen atom, a hydroxyl group or an alkoxy group with 1-5 carbon atoms; precursors capable of liberating a substance according to the aforementioned formula when used in the present method; and mixtures of one or more of the aforementioned substances and / or precursors. The estrogenic component according to the invention is particularly useful in the treatment or prevention of colorectal and prostate cancer and, unlike commonly used estrogens, does not simultaneously enhance the risk of estrogen-stimulated cancers such as breast cancer.
Owner:ESTETRA SRL
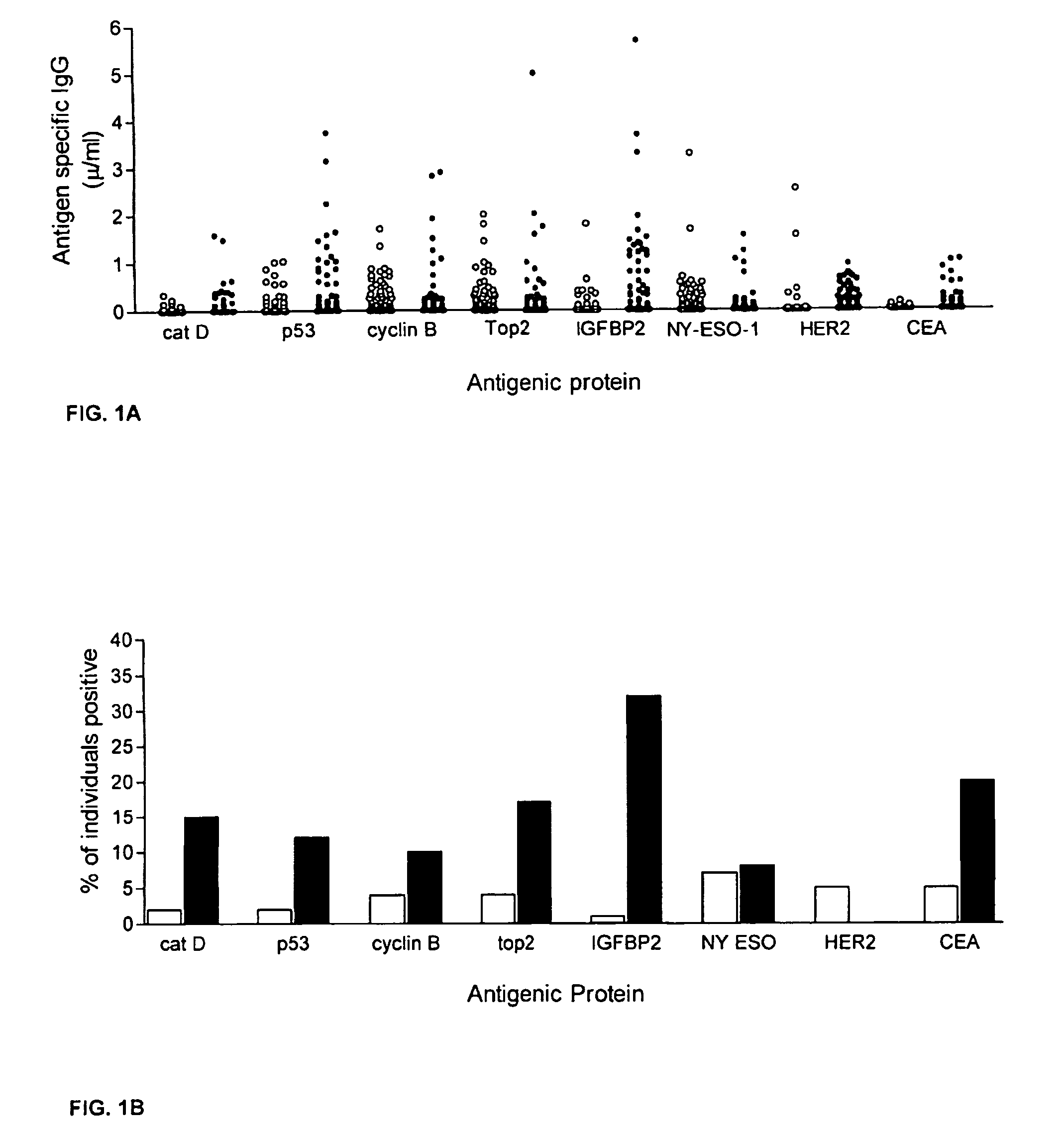
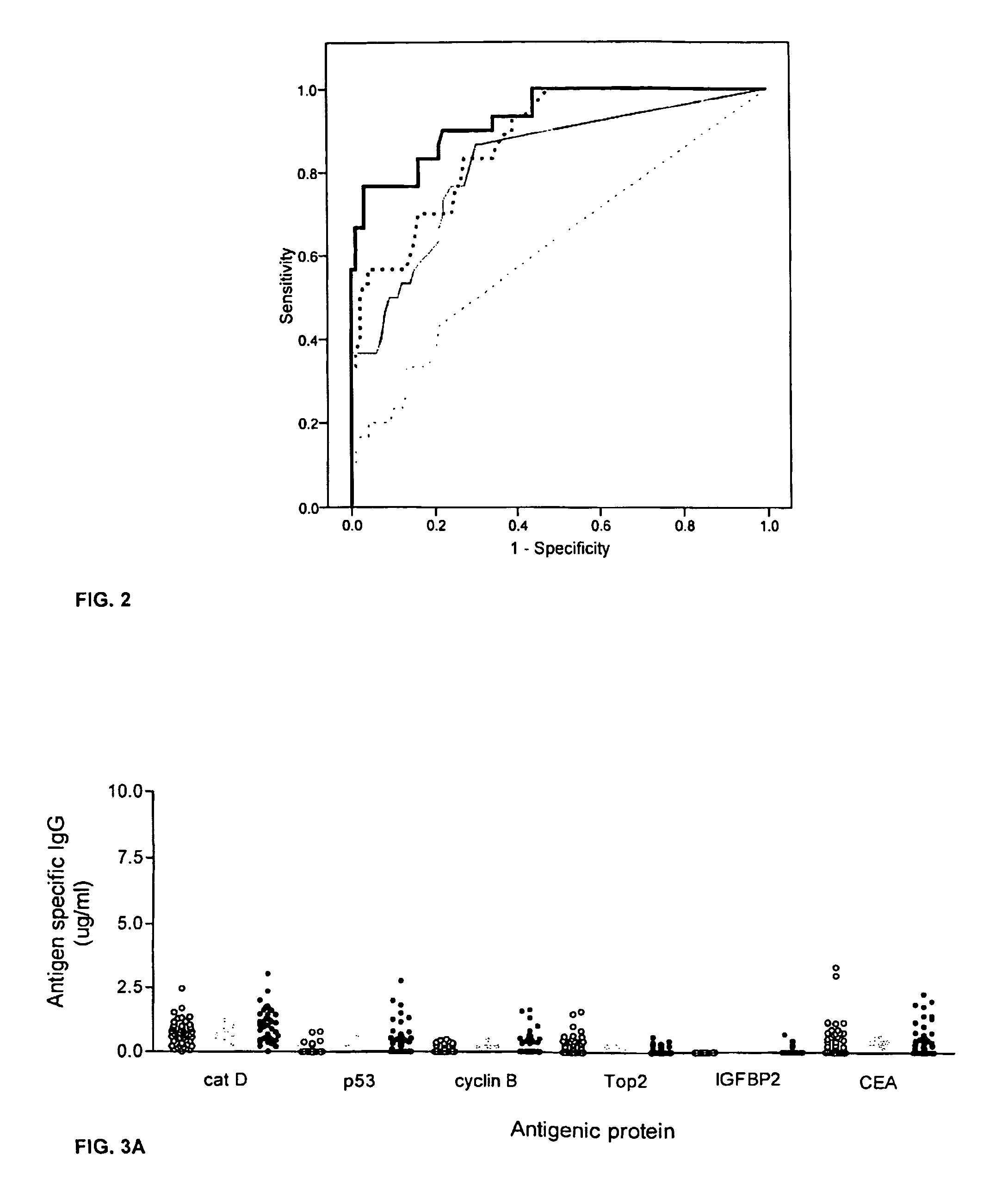
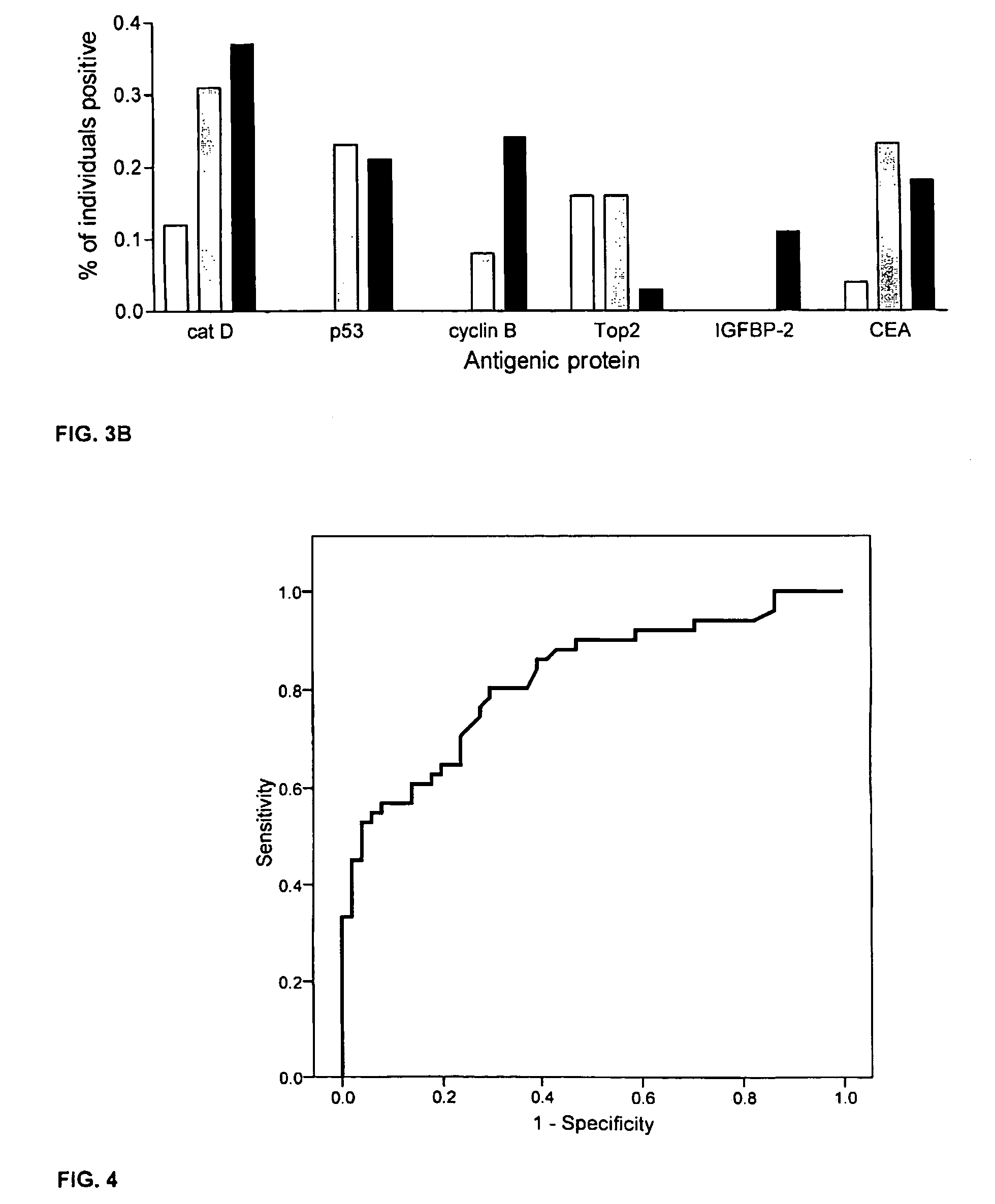
Diagnostic panel of cancer antibodies and methods for use
The invention provides a method for detection of a malignancy in a specimen of bodily fluid. The method comprises contacting the specimen with at least two antigens selected from the group consisting of p53, IGFBP2, Topo2α, cathepsin D, cyclin B, cyclin D1, MUC1, HER-2 / neu and CEA. The method further comprises incubating the specimen and the antigen for a duration and under conditions that are sufficient for the formation of immunocomplexes; and detecting the presence or absence of immunocomplex formation between the antigens and antibodies specific for the antigens in the specimen, thereby determining the presence or absence of the malignancy. Also provided is a method for monitoring the effectiveness of cancer therapy related to a malignancy in a warm-blooded animal, a method for distinguishing between Stage I and Stage II colorectal cancer in a specimen of bodily fluid.
Owner:UNIV OF WASHINGTON
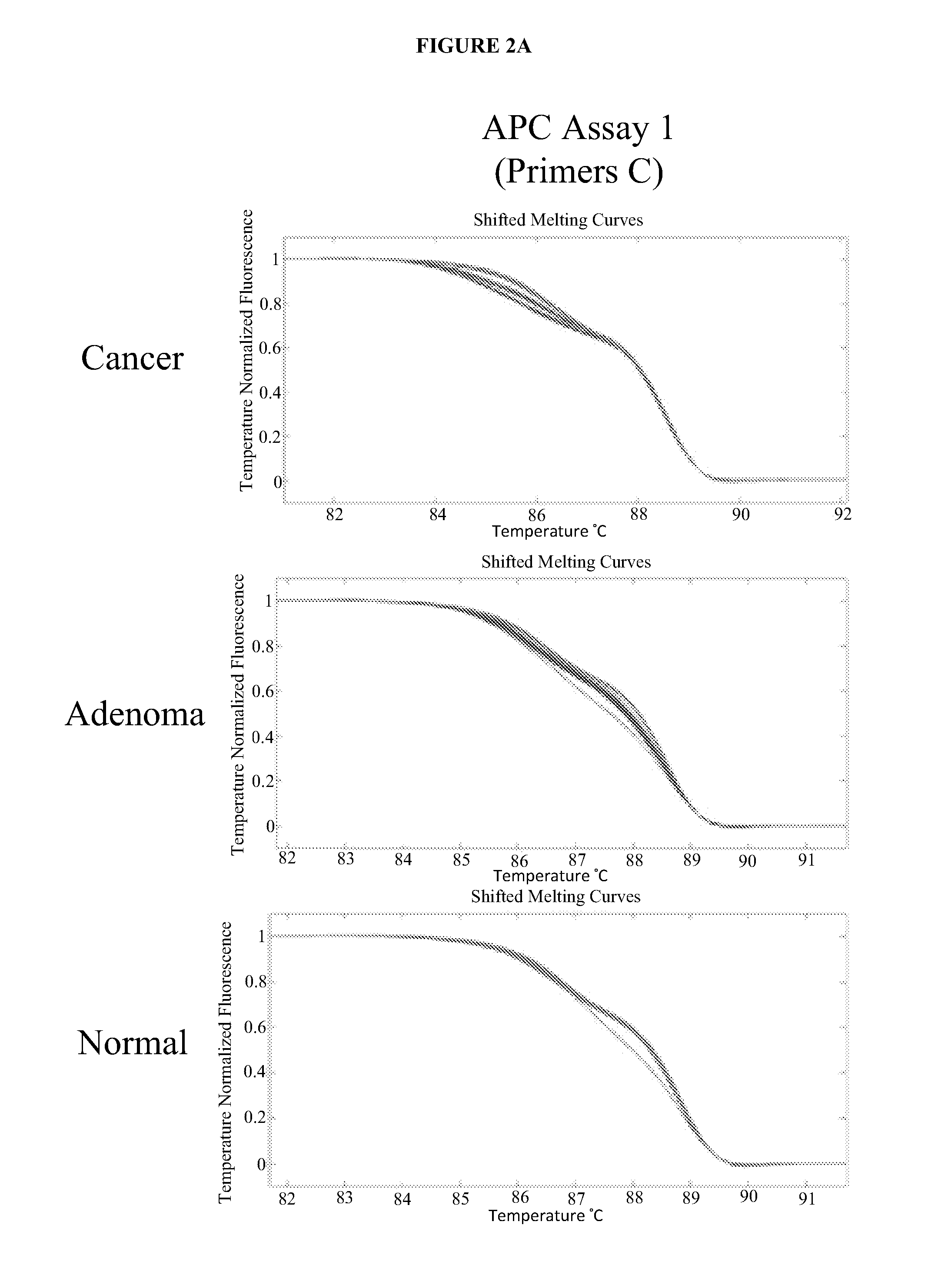
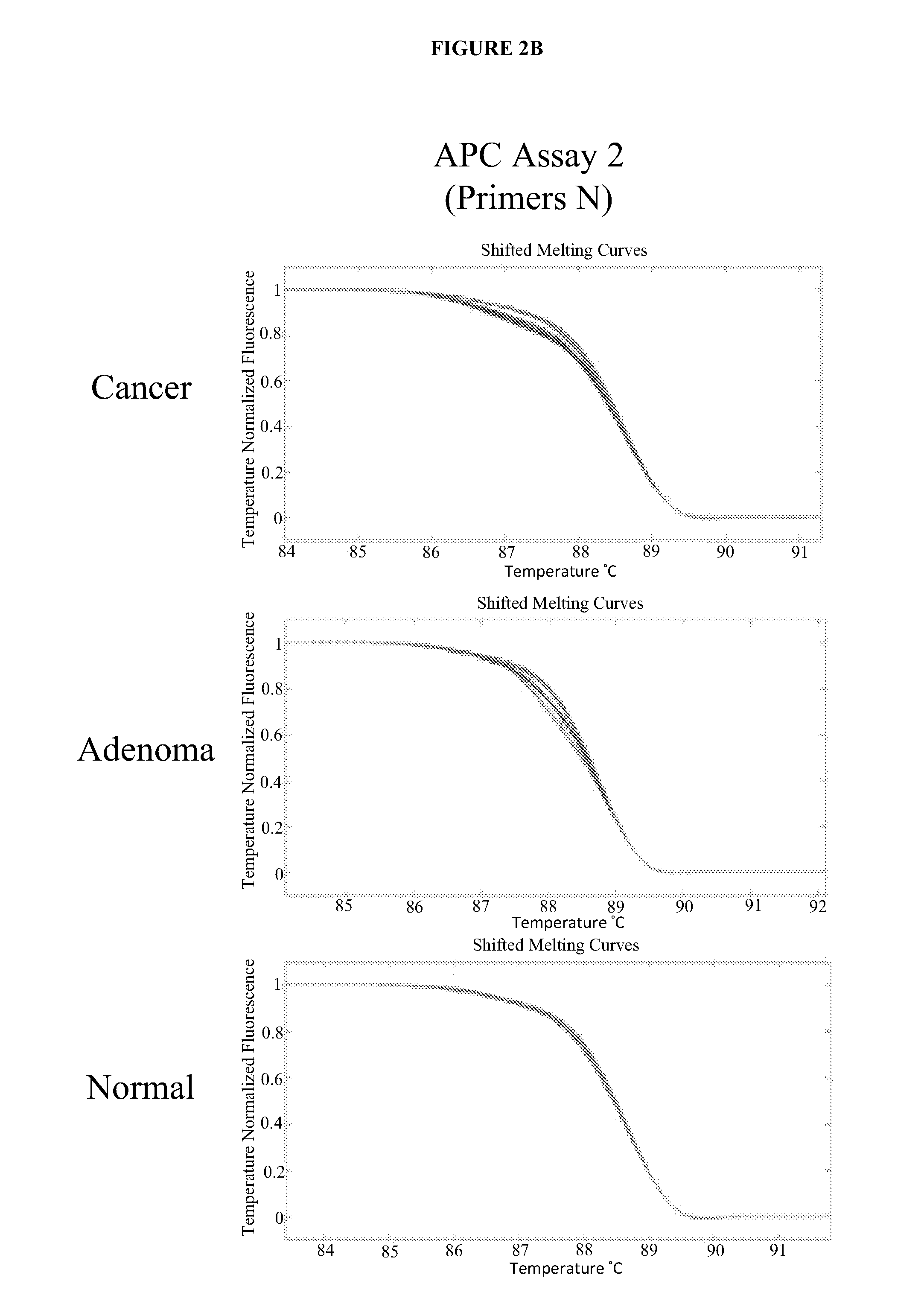
Methods and materials for detecting colorectal cancer and adenoma
InactiveUS20130012410A1High detectionHigh level of sensitivityMicrobiological testing/measurementLibrary screeningFOXE1Mammal
The present invention provides methods and materials related to the detection of colorectal neoplasm-specific markers (e.g., markers associated with colorectal cancer, markers associated with adenoma) in or associated with a subject's stool sample. In particular, the present invention provides methods and materials for identifying mammals (e.g., humans) having a colorectal neoplasm by detecting the presence and level of indicators of colorectal neoplasia such as, for example, long DNA (e.g., quantified by Alu PCR) and the presence and level of tumor-associated gene alterations (e.g., mutations in KRAS, APC, melanoma antigen gene, p53, BRAF, BAT26, PIK3CA) or epigenetic alterations (e.g., DNA methylation) (e.g., CpG methylation) (e.g., CpG methylation in coding or regulatory regions of bmp-3, bmp-4, SFRP2, vimentin, septin9, ALX4, EYA4, TFPI2, NDRG4, FOXE1) in DNA from a stool sample obtained from the mammal.
Owner:MAYO FOUND FOR MEDICAL EDUCATION & RES
Markers for colorectal cancer
InactiveUS20060188883A1Reduce and eliminate biological activityReduce expressionMicrobiological testing/measurementDisease diagnosisCancer cellBifunctional
Provided are previously uncharacterised markers of cancers, for example colorectal cancers, and uses of these as diagnostic and prognostic markers of cancers, and in particular colorectal cancers. The markers are SEQ ID NO: 1—hnRNP-K; SEQ ID NO:2—HMG-1; SEQ ID NO:3—proteasome subunit alpha type 1; SEQ ID NO:4—bifunctional purine biosynthesis protein; SEQ ID NO:5—ST11; SEQ ID NO:6—annex in IV; SEQ ID NO:7—60 kDa heat shock protein; SEQ ID NO:8—T complex protein 1 beta subunit; SEQ ID NO:9—T complex protein 1 epsilon subunit; SEQ ID NO: 10—mortalin; and SEQ ID NO: 11—TER-ATPase. The invention further provides related methods and materials for the use of the markers in therapeutic intervention in colorectal and other cancers e.g. to specifically target neoplastic cells without causing significant toxicity in healthy tissues, and to provide methods for the evaluation of the ability of candidate therapeutic compounds to modulate the biological activity of cancerous cells from the colon, rectum and other tissues.
Owner:AUVATION +2
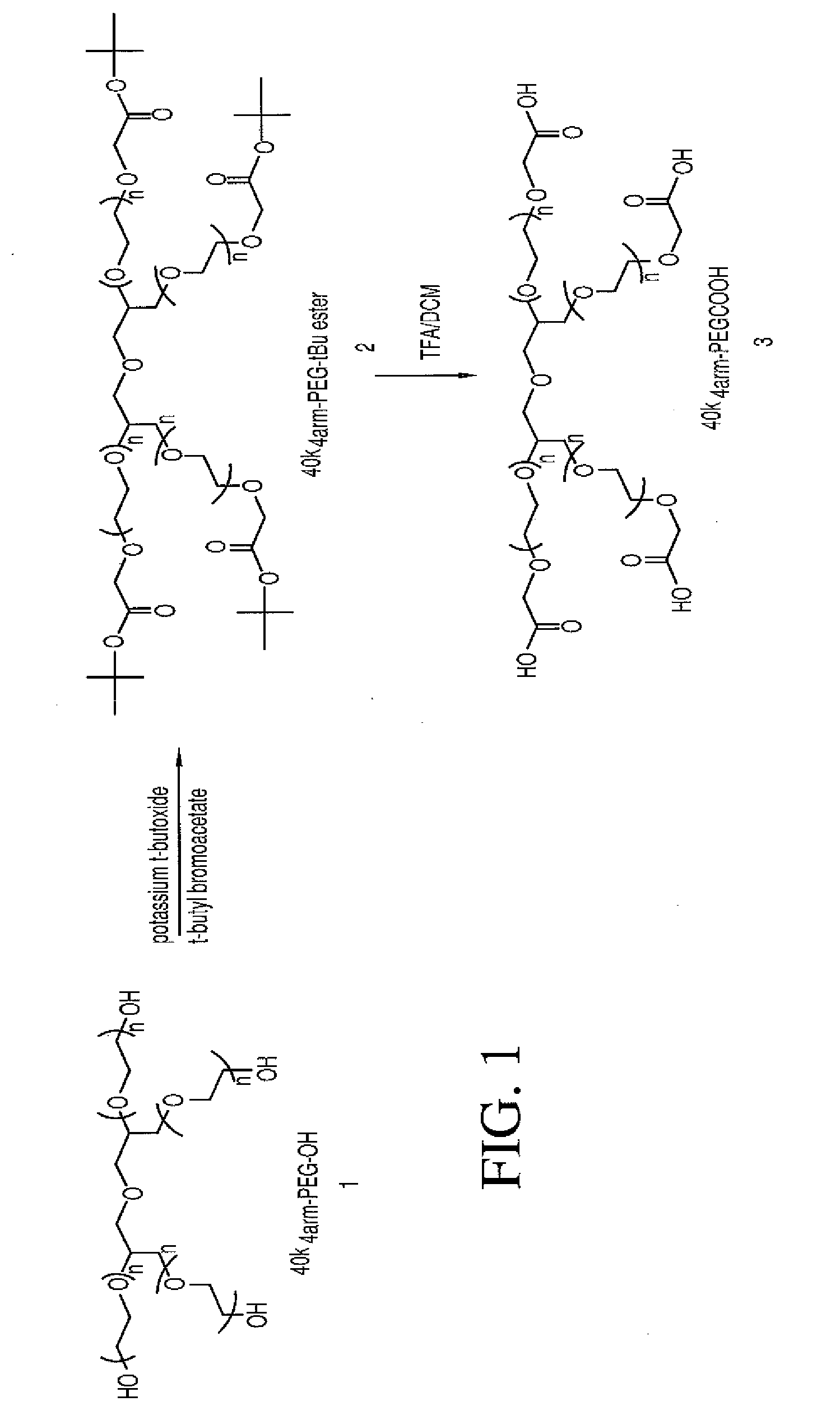
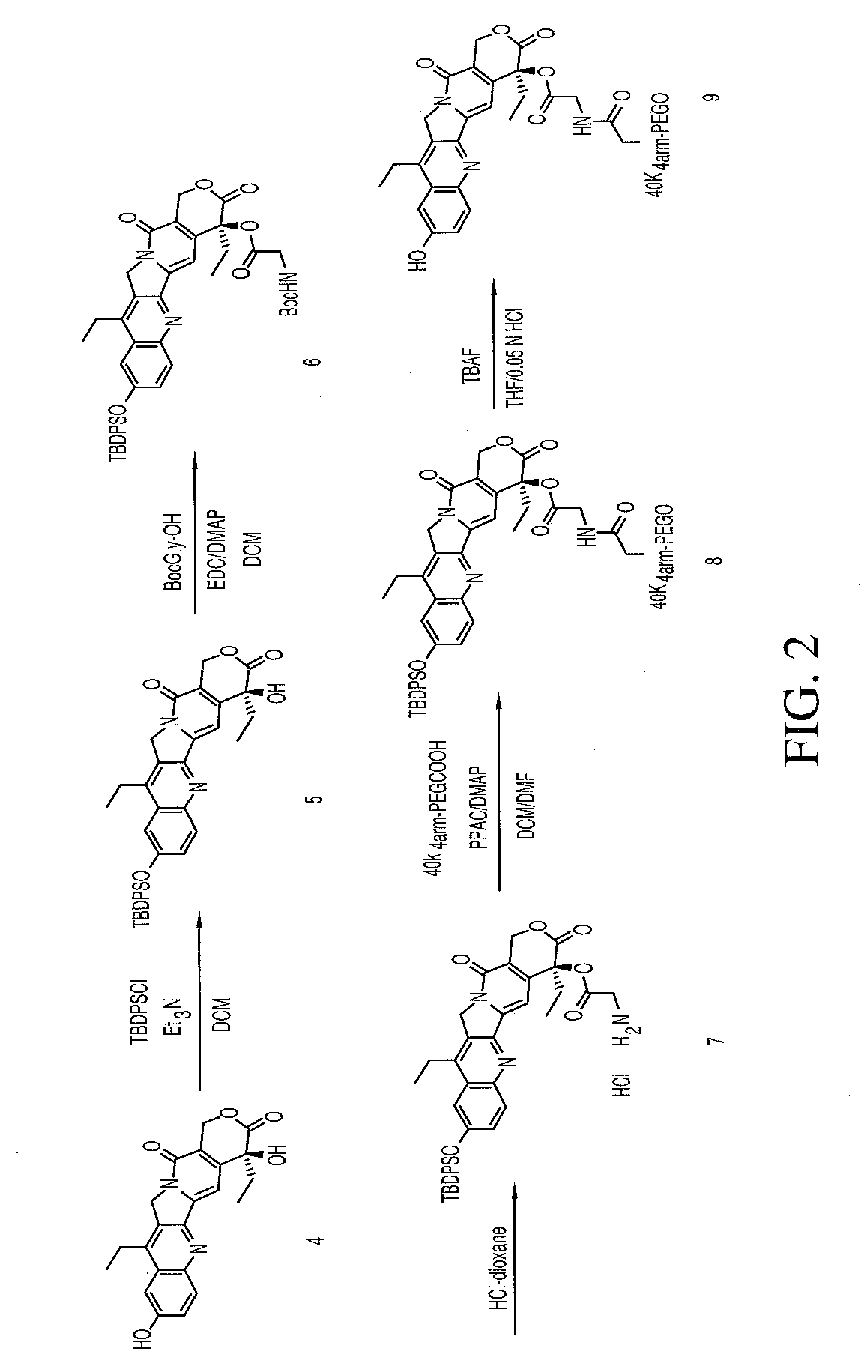
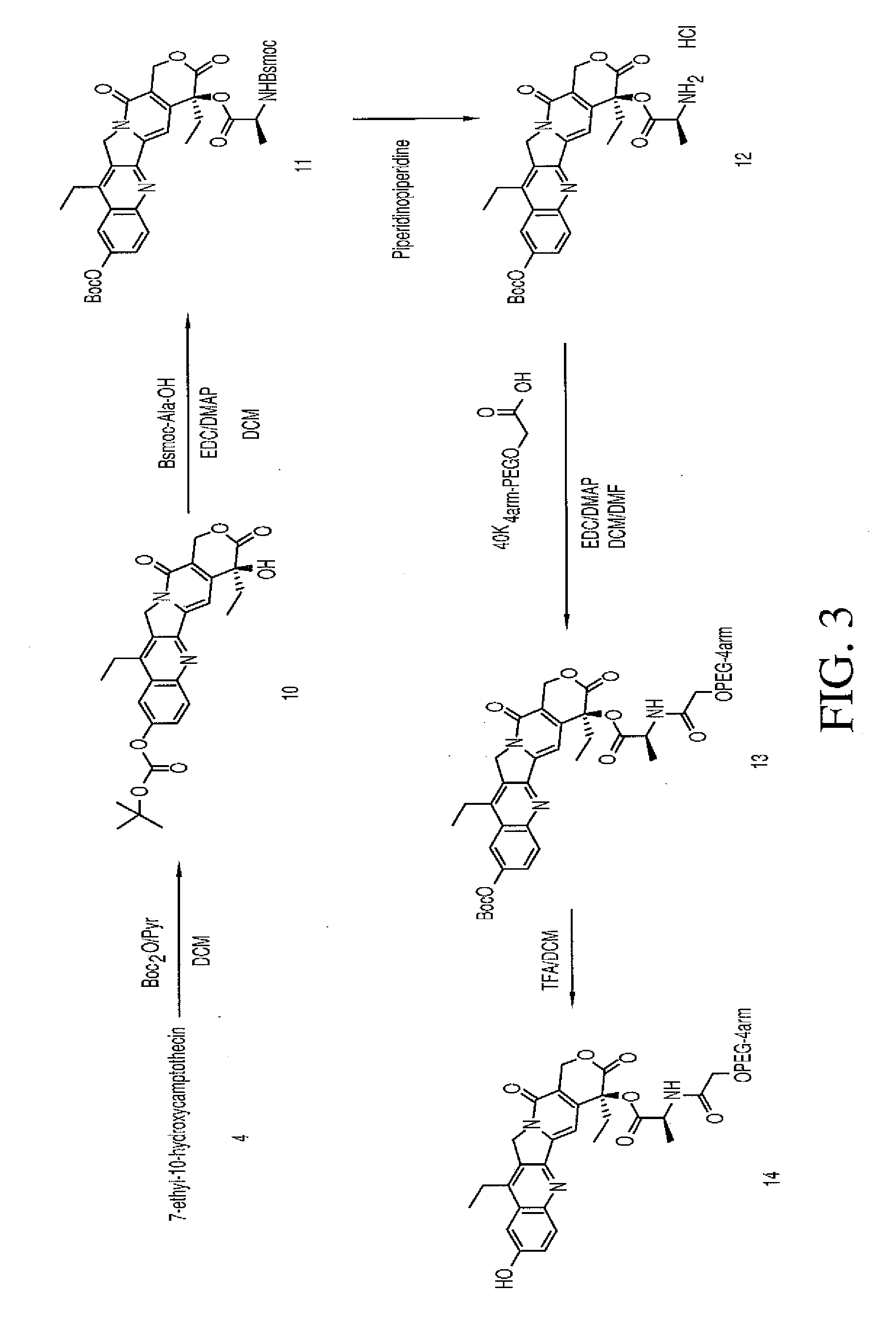
Multi-arm polymeric conjugates of 7-ethyl-10-hydroxycamptothecin for treatment of breast, colorectal, pancreatic, ovarian and lung cancers
InactiveUS20070197575A1Advanced technologyGood conjugateBiocideOrganic chemistryMedicinePolyethylene glycol
A four arm-polyethylene glycol-7-ethyl-10-hydroxycamptothecin conjugate, such as, is disclosed. Methods of making the conjugates and methods of treating mammals using the same are also disclosed.
Owner:BELROSE PHARMA
Biomarkers
InactiveUS20150141273A1Conducive to screeningUseful in detectionCompound screeningApoptosis detectionCvd riskBiomarker (petroleum)
The invention provides a method for screening for colorectal cancer, the method comprising: screening a biological sample from an individual for one or more biomarkers selected from the group defined in Table 1 and / or Table 6, wherein the presence of or increased expression of the one or more biomarkers relative to a control sample is indicative that the individual is at risk of suffering from or is suffering from colorectal cancer. The invention also provides an array and kit suitable for use in the methods of the invention, methods of treating colorectal cancer and therapeutic agents for use in methods of treating cancer.
Owner:STICHTING VU VUMC
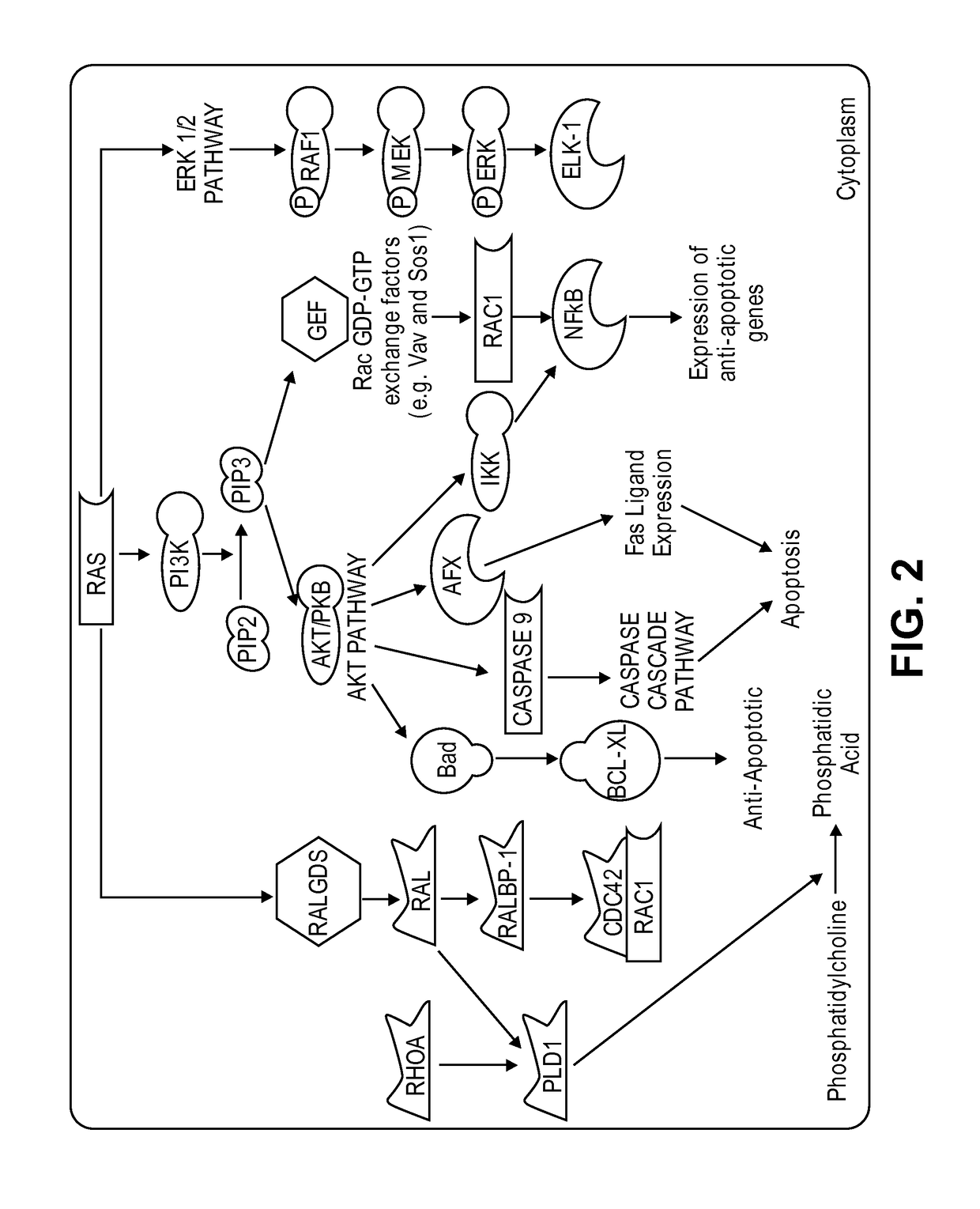
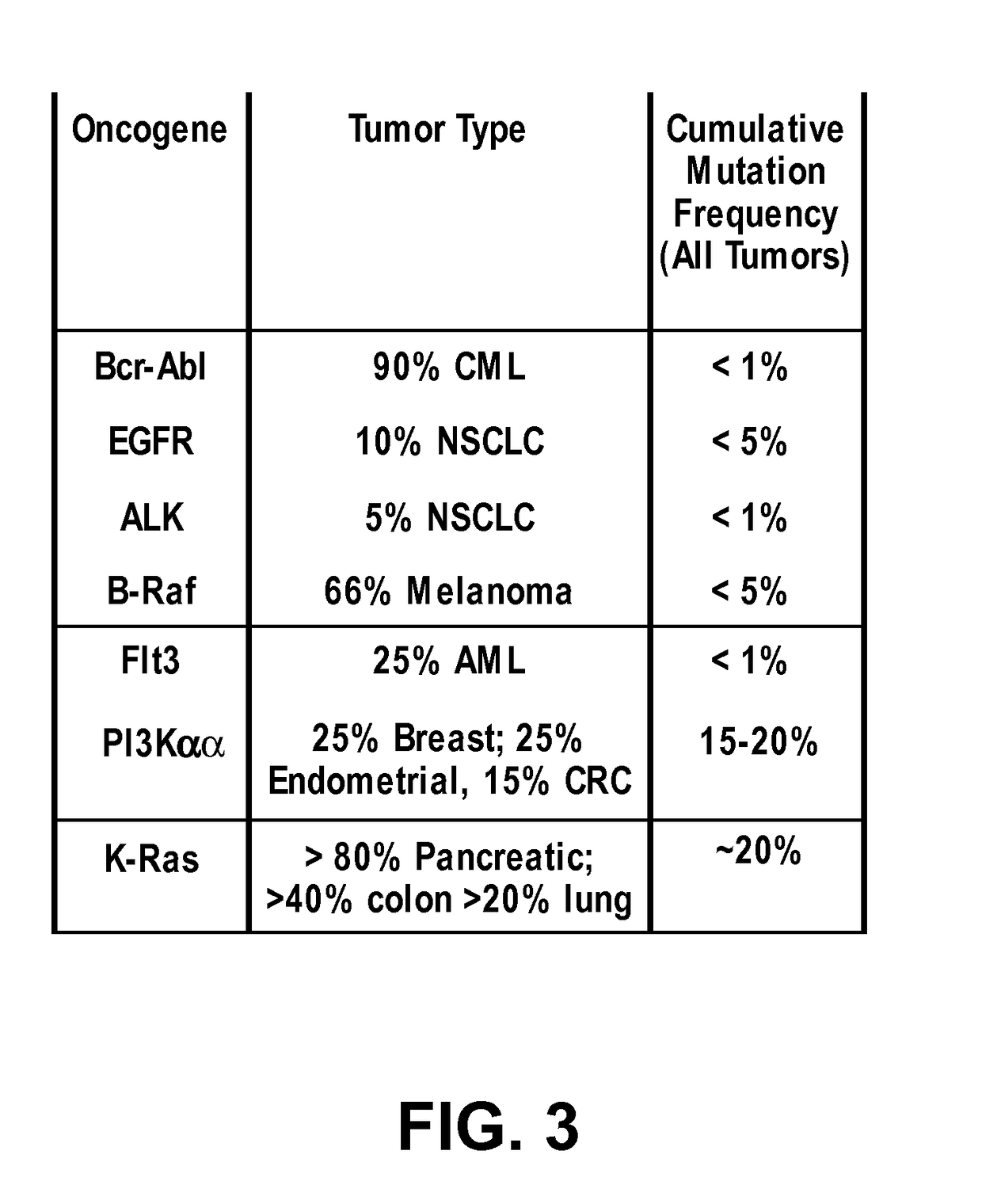
Methods and compositions for inhibition of ras
Inhibitors of Ras protein, methods to modulate the activity of Ras protein, and methods of treatment of disorders mediated by Ras protein are provided. A method for regulating activity of a K-Ras, H-Ras or N-Ras mutant protein with a compound is described. Disorders that can be treated include cancer, such as hematological cancer, pancreatic cancer, MYH associated polyposis, colorectal cancer, or lung cancer.
Owner:ARAXES PHARMA LLC
Methods of detecting colorectal cancer
InactiveUS20120164238A1Prevents denaturation and degradationBiocideInorganic active ingredientsColorectal cancerOncology
A method of detecting a predisposition to, or the incidence of colorectal cancer in a faecal sample comprises, in a first step (a), detecting the presence of blood in the faecal sample, wherein detection of the presence of blood is indicative of a predisposition to, or the incidence of colorectal cancer. The method additionally comprises, in second step (b), detecting an epi-genetic modification in the DNA contained within the faecal sample, wherein detection of the epigenetic modification is indicative of a predisposition to, or the incidence of colorectal cancer. Based upon a positive result obtained in either (a) or (b) or in both (a) and (b) a predisposition to, or the incidence of colorectal cancer is detected. Related methods and kits involve detecting an epigenetic modification in a number of specific genes.
Owner:EXACT SCI CORP
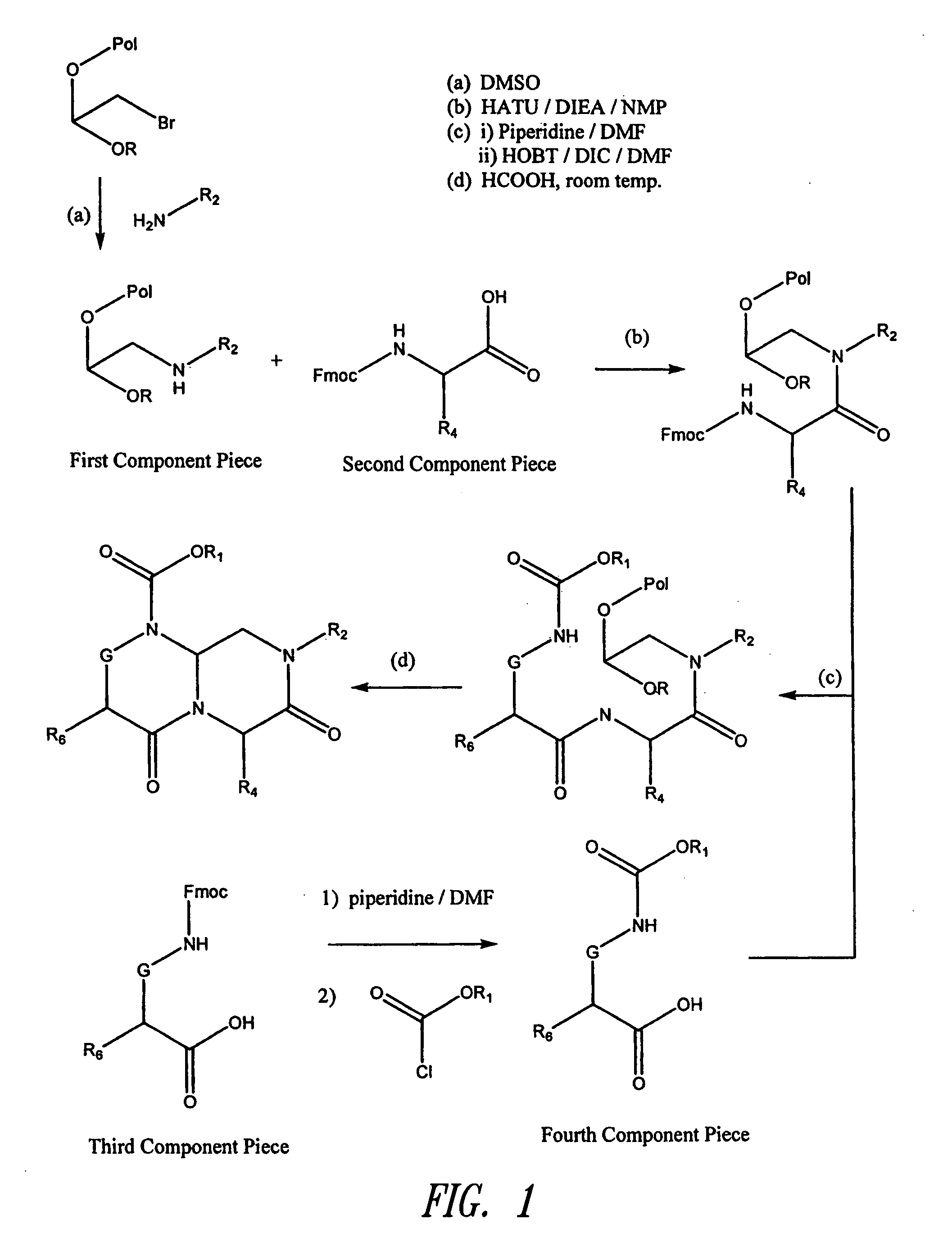
Reverse-turn mimetics and method relating thereto
InactiveUS20070021425A1Efficient implementationBiocideNervous disorderUlcerative colitisPercent Diameter Stenosis
Conformationally constrained compounds that mimic the secondary structure of reverse-turn regions of biologically active peptides and proteins are disclosed. Such reverse-turn mimetic structures have utility over a wide range of fields, including use as diagnostic and therapeutic agents. Libraries containing the reverse-turn mimetic structures of this invention are also disclosed as well as methods for screening the same to identify biologically active members. The invention also relates to the use of such compounds for inhibiting or treating disorders modulated by Wnt-signaling pathway, such as cancer, especially colorectal cancer, restenosis associated with angioplasty, polycystic kidney disease, aberrant angiogenesis disease, rheumatoid arthritis disease, tuberous sclerosis complex, Alzheimer's disease, excess hair growth or loss, or ulcerative colitis.
Owner:JW PHARMA CORP
Reverse-turn mimetics and method relating thereto
Conformationally constrained compounds that mimic the secondary structure of reverse-turn regions of biologically active peptides and proteins are disclosed. Such reverse-turn mimetic structures have utility over a wide range of fields, including use as diagnostic and therapeutic agents. Libraries containing the reverse-turn mimetic structures of this invention are also disclosed as well as methods for screening the same to identify biologically active members. The invention also relates to the use of such compounds for inhibiting or treating disorders modulated by Wnt-signaling pathway, such as cancer, especially colorectal cancer, restenosis associated with angioplasty, polycystic kidney disease, aberrant angiogenesis disease, rheumatoid arthritis disease, tuberous sclerosis complex, Alzheimer's disease, excess hair growth or loss, or ulcerative colitis.
Owner:JW PHARMA CORP
Kinase inhibitors useful for the treatment of proliferative diseases
The present invention relates to novel kinase inhibitors and modulator compounds useful for the treatment of various diseases. More particularly, the invention is concerned with such compounds, kinase / compound adducts, methods of treating diseases, and methods of synthesis of the compounds. Preferrably, the compounds are useful for the modulation of kinase activity of Raf kinases and disease polymorphs thereof. Compounds of the present invention find utility in the treatment of mammalian cancers and especially human cancers including but not limited to malignant melanoma, colorectal cancer, ovarian cancer, papillary thyroid carcinoma, non small cell lung cancer, and mesothelioma. Compounds of the present invention also find utility in the treatment of rheumatoid arthritis and retinopathies including diabetic retinal neuropathy and macular degeneration.
Owner:DECIPHERA PHARMA LLC
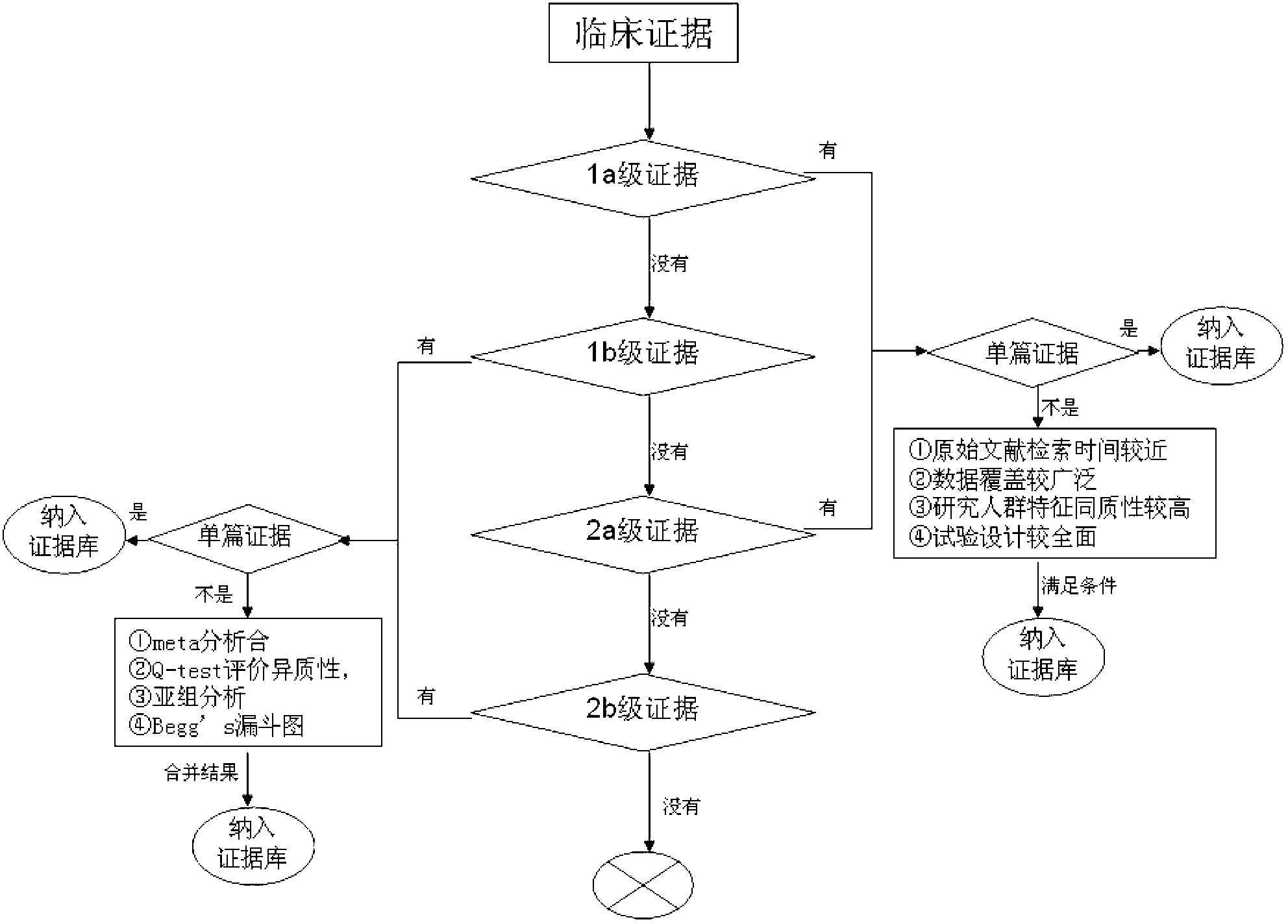
Screening method of colorectal cancer treatment prognosis biomarkers
InactiveCN103324846AShorten screening timeReduce screening costsSpecial data processing applicationsDNA/RNA fragmentationKRASPrognosis biomarker
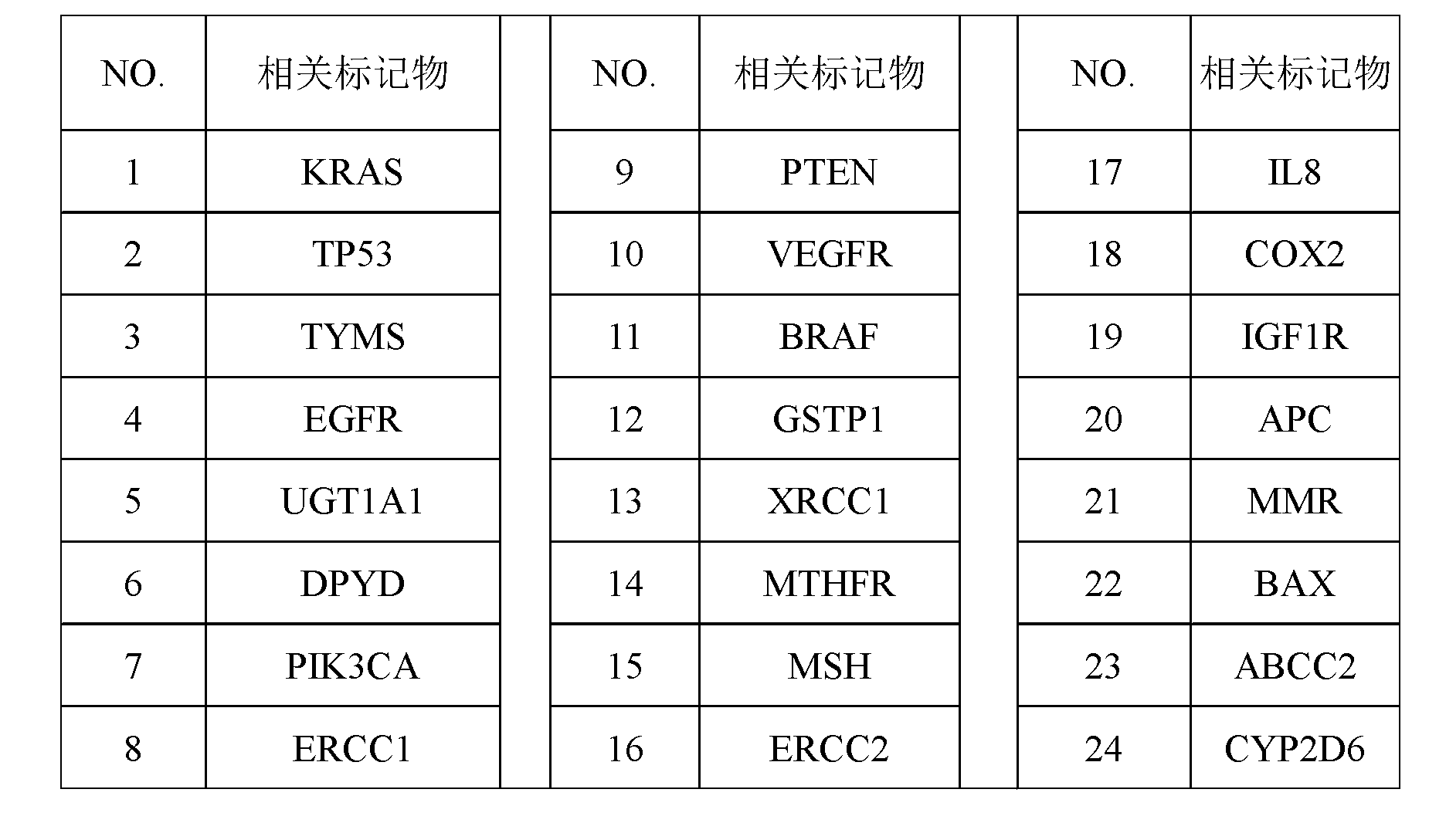
The invention discloses a screening method of colorectal cancer treatment prognosis biomarkers. The screening method includes the following steps of: (1) making a primary search strategy for screening the biomarkers; (2) making a literature adopting and exclusion criterion; (3) analyzing data and primarily screening the biomarkers; (4) making evident grades; (5) making a high quality evident evaluation criterion; (6) performing grading retrieval and quality evaluation on biomarker clinic evident; (7) performing data analyzing and statistics on the biomarker clinic evident; (8) screening the prognosis biomarkers. The invention further provides the colorectal cancer treatment prognosis biomarkers obtained through screening according to the screening method, and the prognosis biomarkers include KRAS, TYMS, TYMS, EGFR, UGT1A1*28, DPYD, PIK3CA, ERCC1, PTEN, VEGFR, BRAF, GST P1, XRCC1, MTHFR, MSI and the like.
Owner:浙江加州国际纳米技术研究院绍兴分院
Method of analysis allowing avoidance of surgery
ActiveUS20170275702A1Prevent removalAvoid surgeryMicrobiological testing/measurementMedical automated diagnosisDNA methylationCervix
A multi-gene-based assay for analysis of the following: (a) oncogene expression; (b) DNA methylation of tumor suppressor genes; (c) non-coding RNA expression (microRNA profiling); and (d) long non-coding RNA expression in cancer samples is disclosed. The assay method and device for conducting the assay are applicable to diagnosis, prognosis, and treatment of various cancers such as lung, breast, colorectal, prostate, liver, bladder; kidney, cervix, pancreatic, gastric, brain, oral, endometrium, and ovary. The assay methods allow avoidance of surgery to remove cancerous tissue.
Owner:GENEVERIFY INC
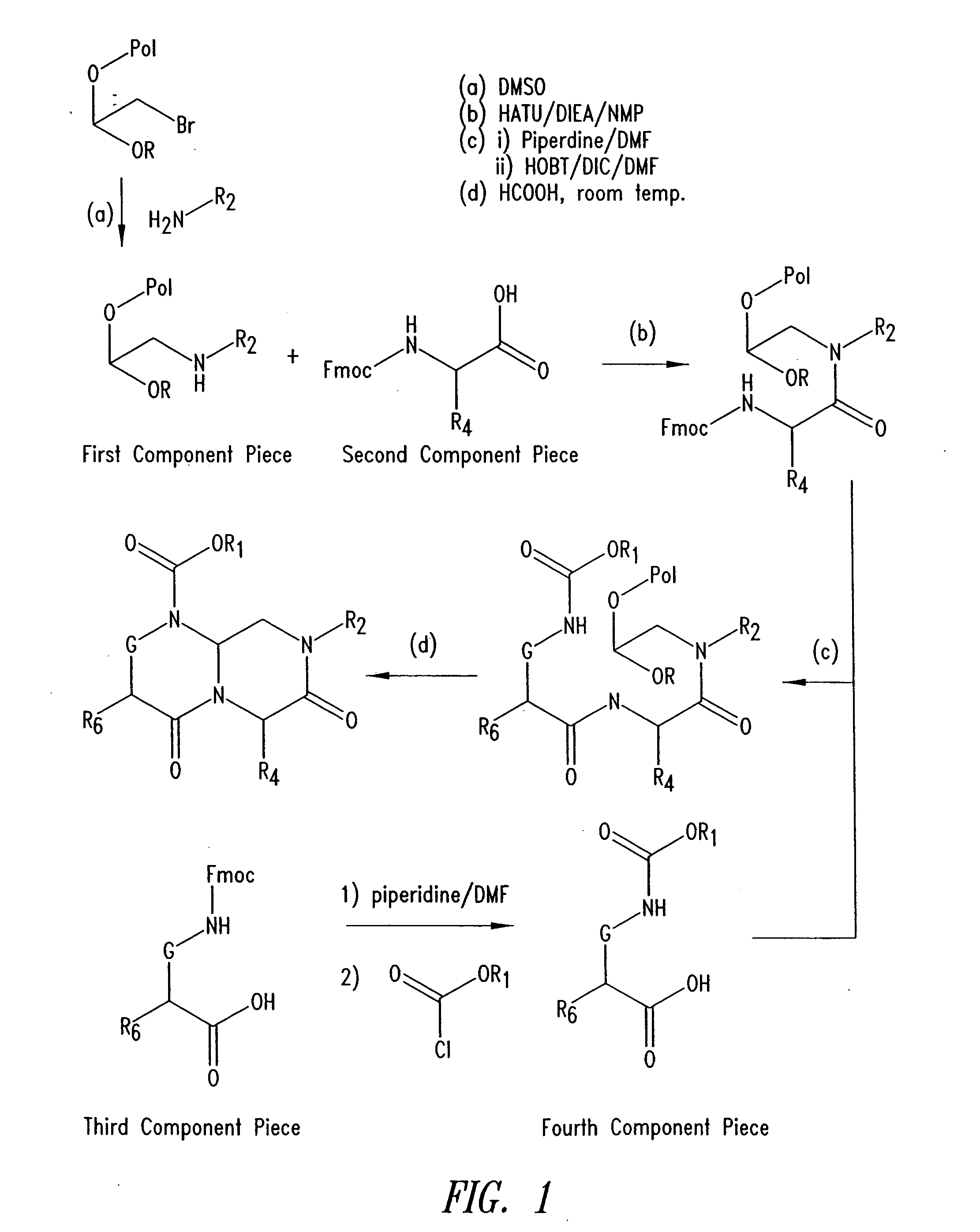
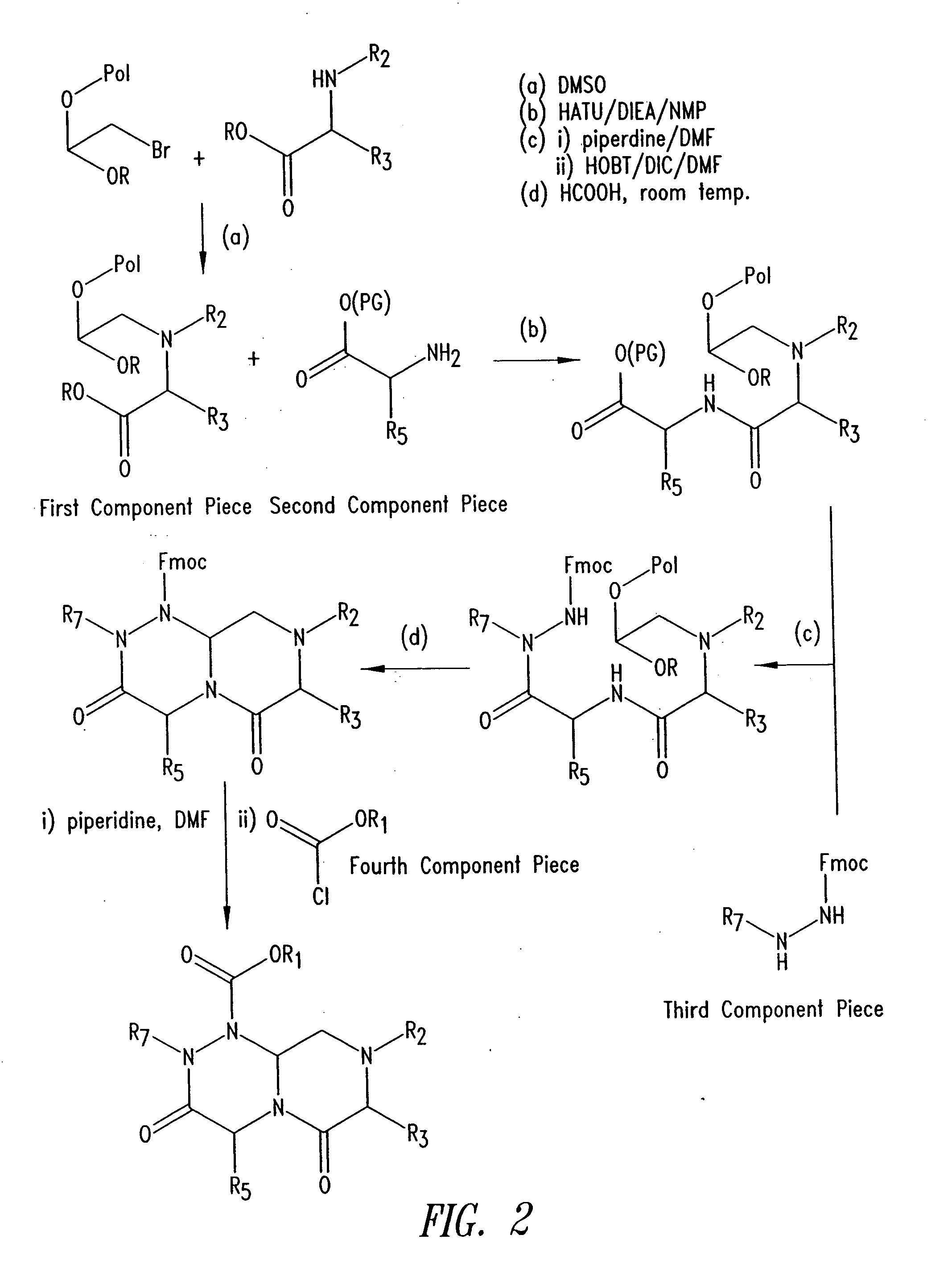
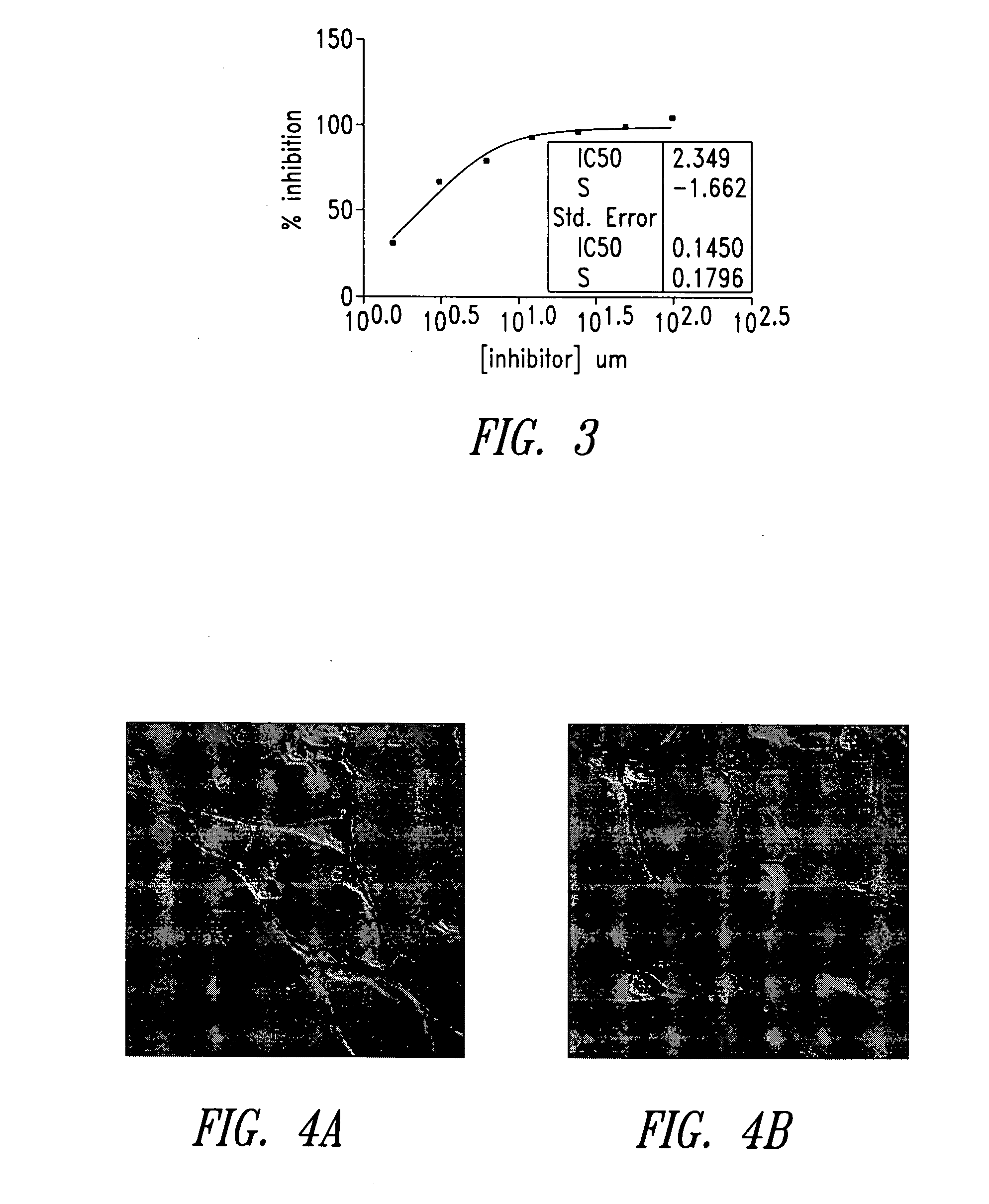
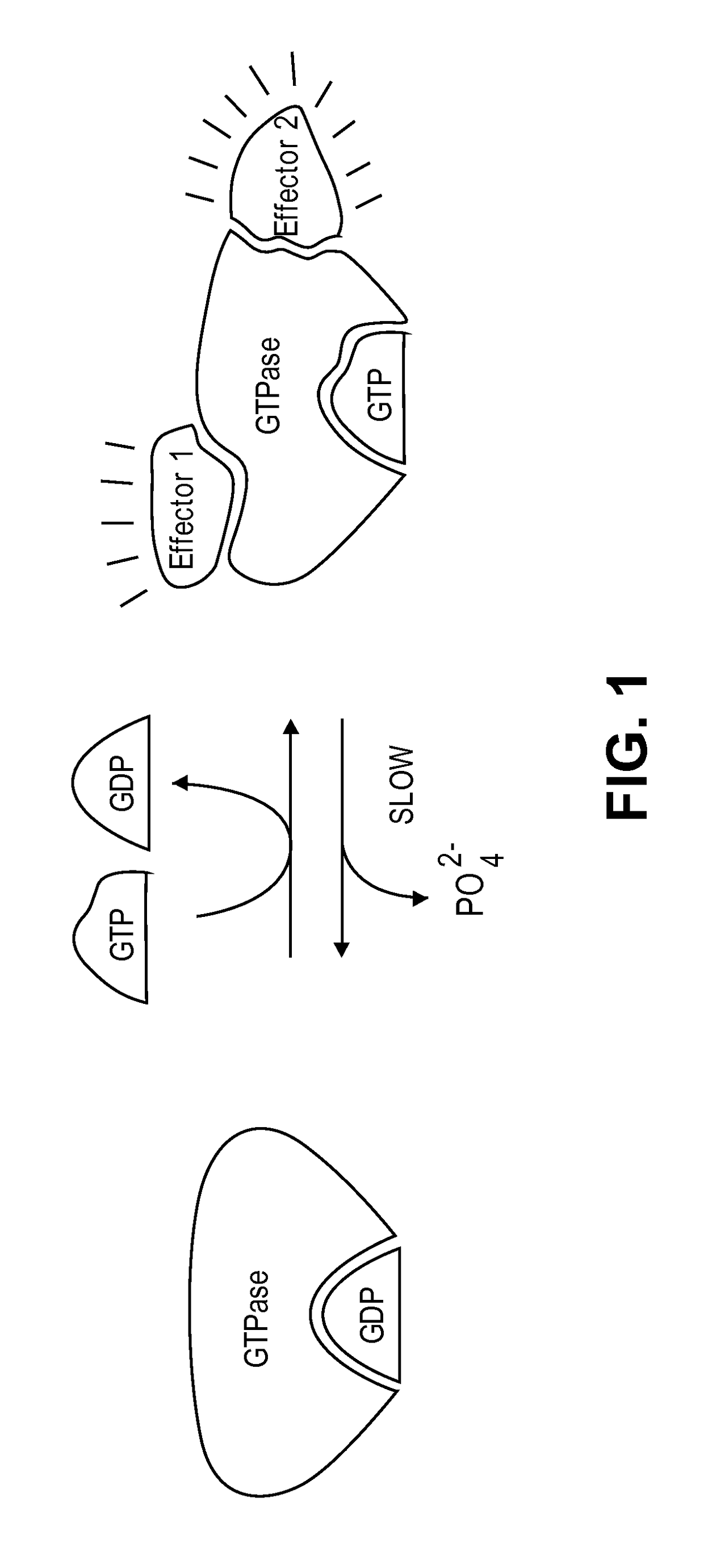
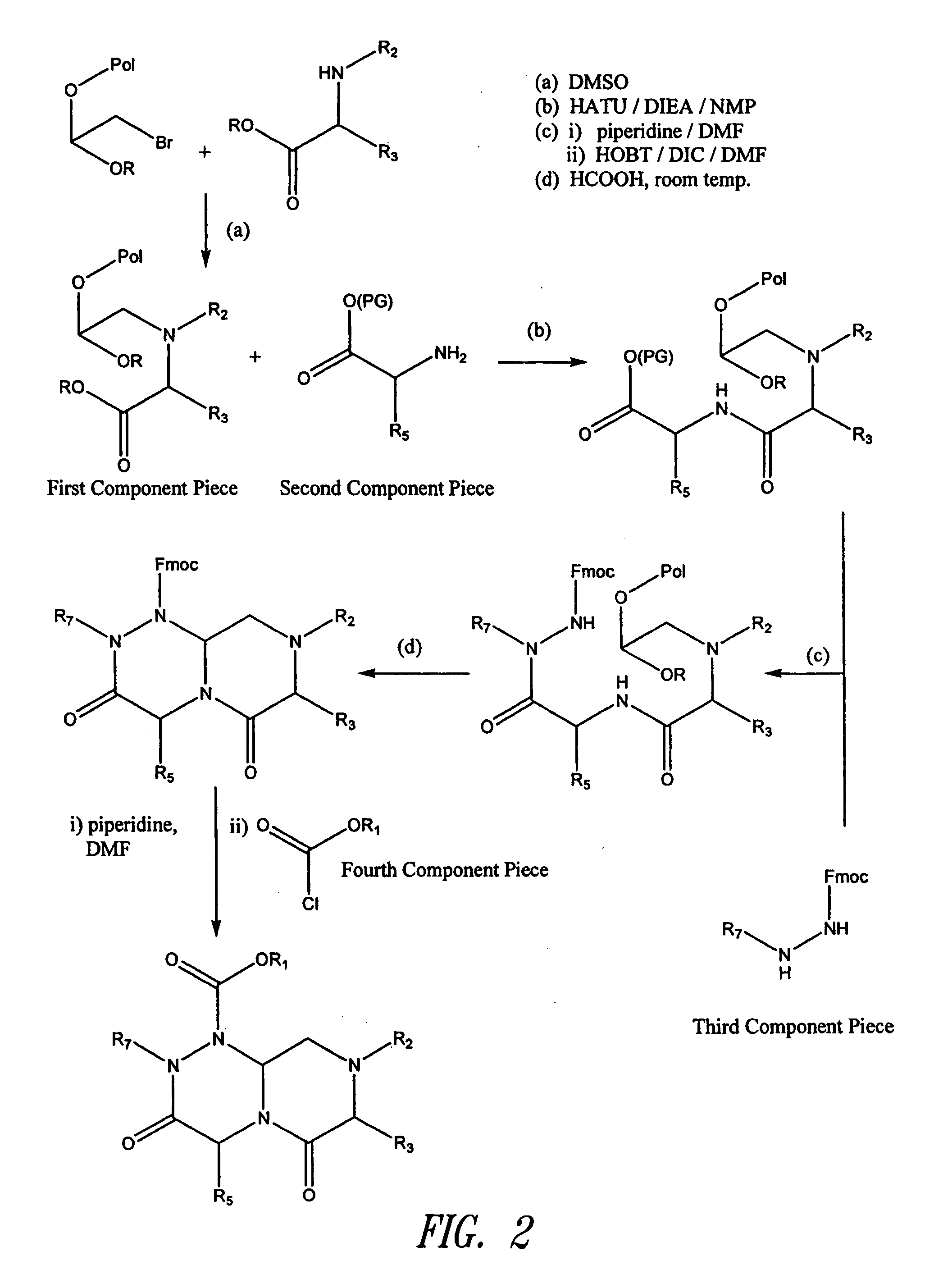
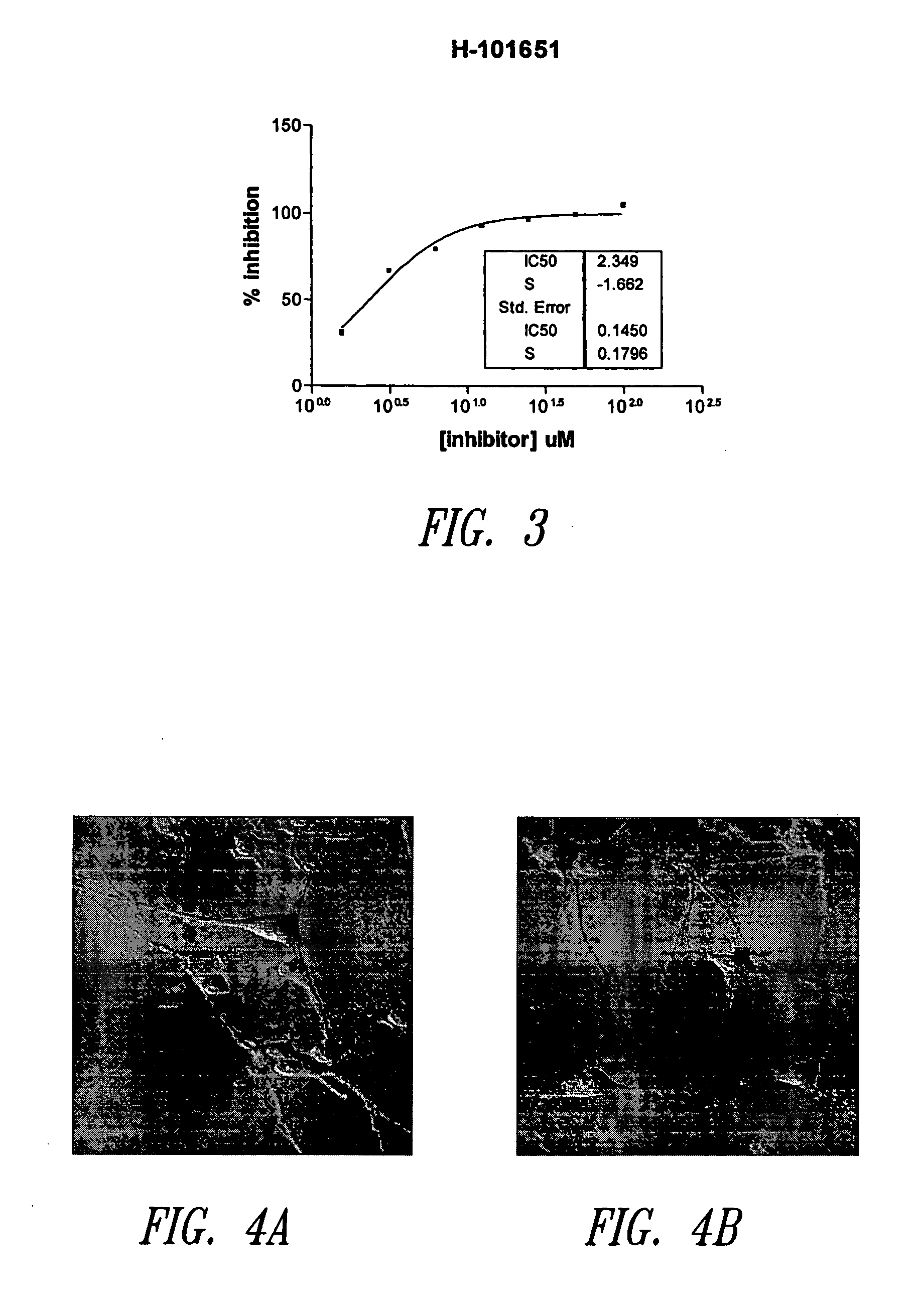
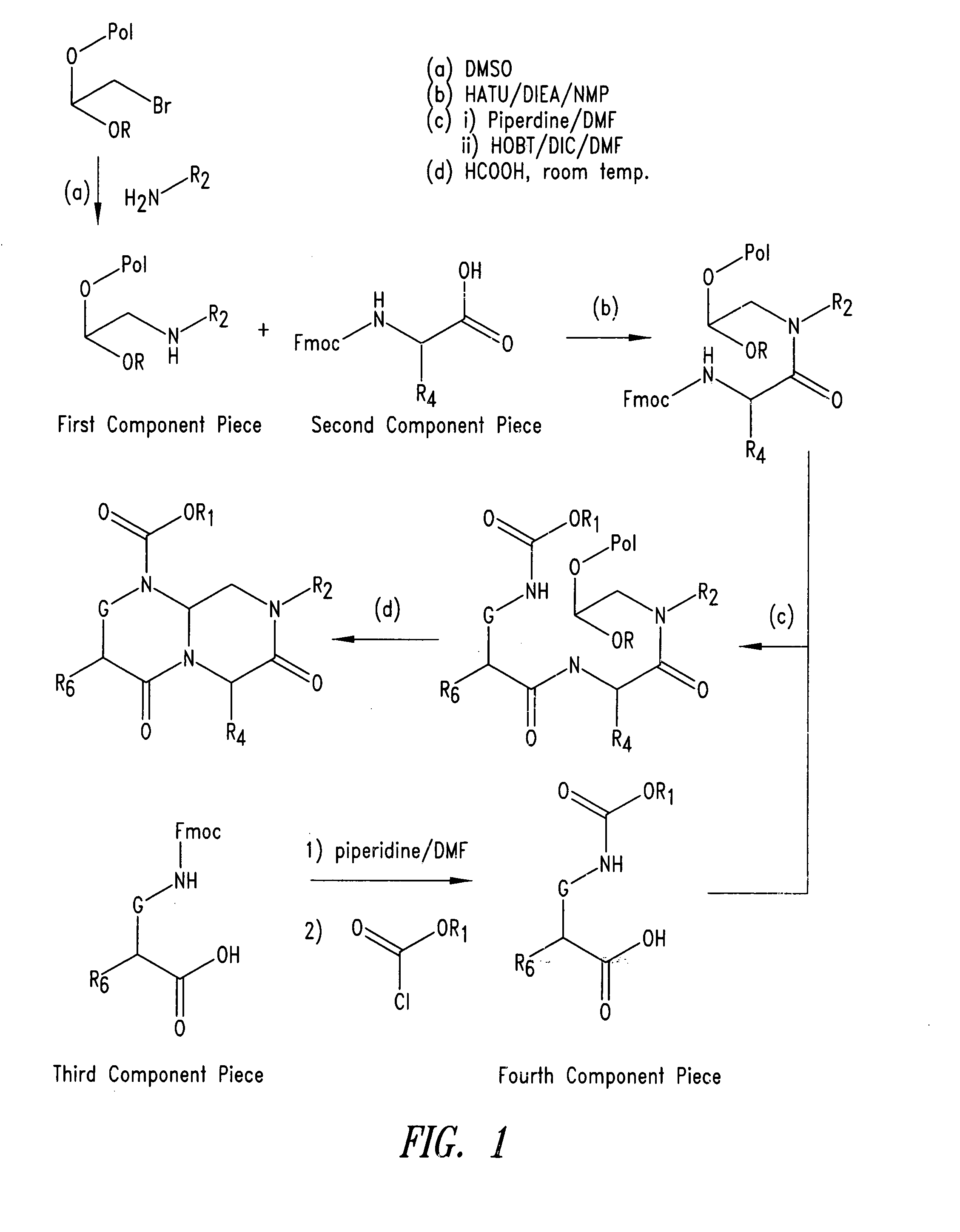
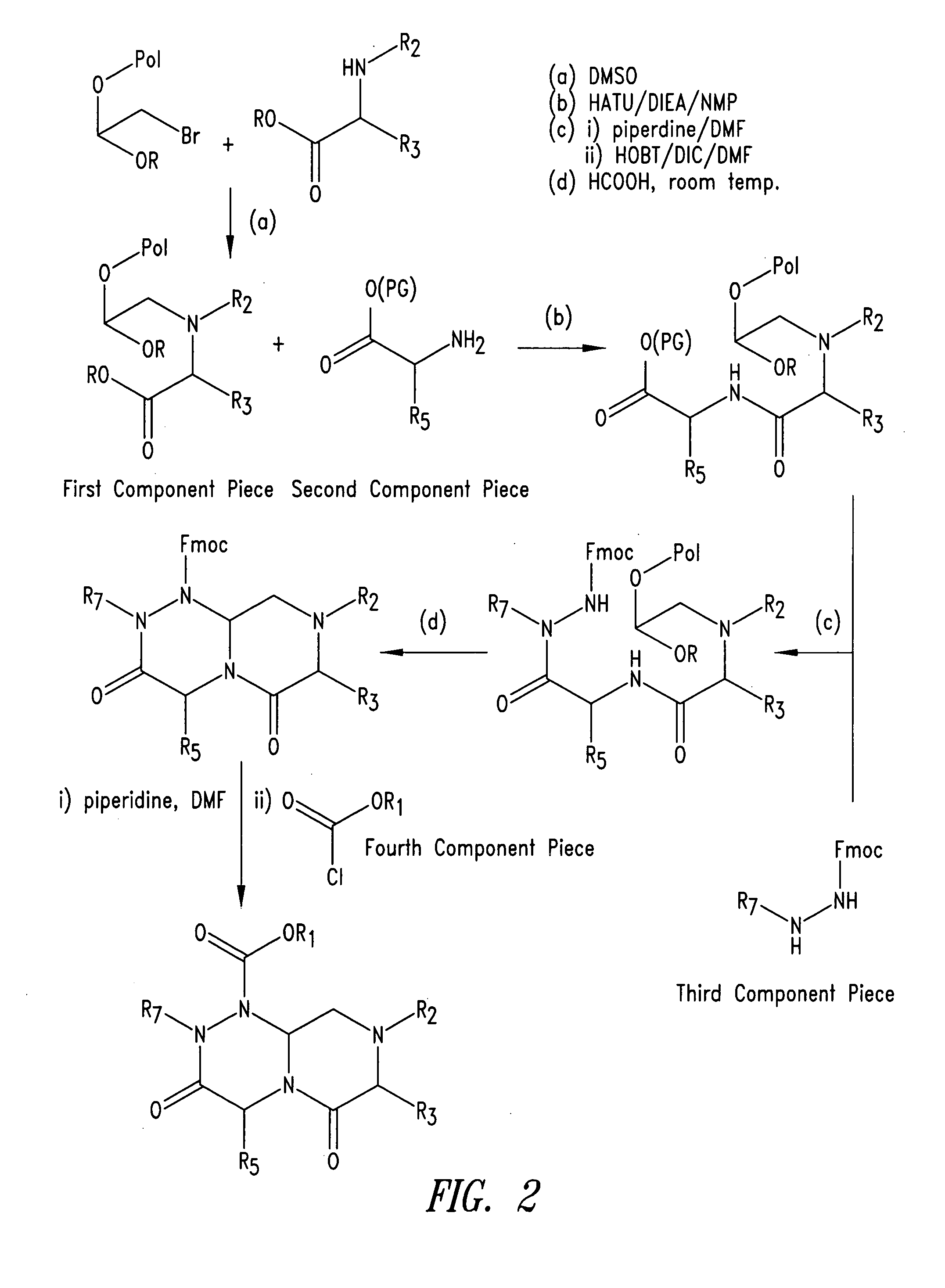
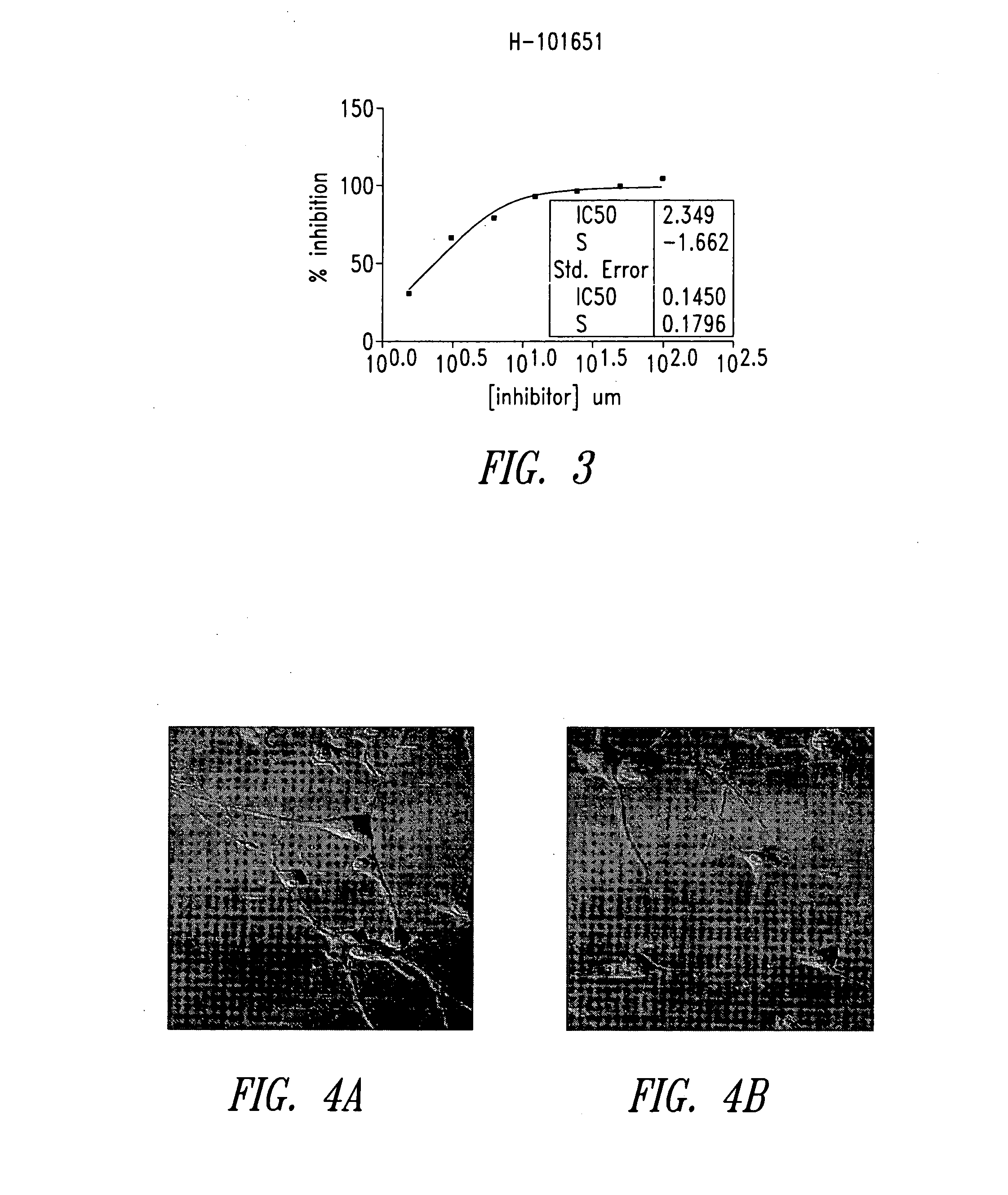
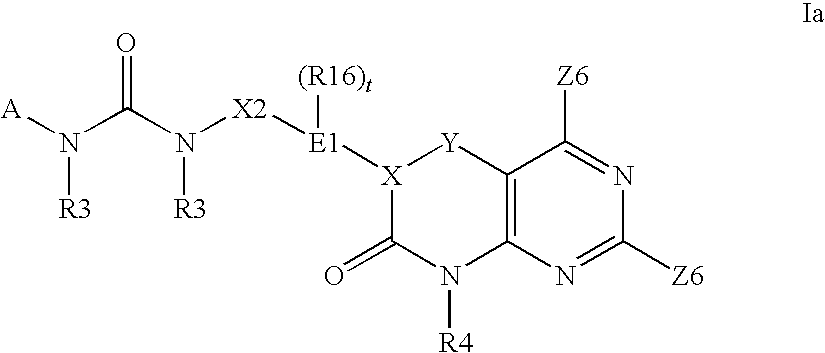
Imidazo[4, 5-B]pyridin-2-one and oxazolo[4, 5-B] pyridin-2-one compounds and analogs thereof as cancer therapeutic compounds
The present invention pertains to certain imidazo[4,5-b]pyridin-2-one and oxazolo[4,5 b]pyridin-2-one compounds and analogs thereof, which, inter alia, inhibit RAF (e.g., B RAF) activity, inhibit cell proliferation, treat cancer, etc. The present invention also pertains to pharmaceutical compositions comprising such compounds, and the use of such compounds and compositions, both in vitro and in vivo, to inhibit RAF (e.g., B-RAF) activity, to inhibit receptor tyrosine kinase (RTK) activity, to inhibit cell proliferation, and in the treatment of diseases and conditions that are ameliorated by the inhibition of RAF, RTK, etc., proliferative conditions such as cancer (e.g., colorectal cancer, melanoma), etc.
Owner:CANCER RES TECH LTD +1
Use and identification of biomarkers for gastrointestinal diseases
InactiveUS20100093552A1Compound screeningApoptosis detectionUlcerative colitisManagement of Crohn's disease
The described invention relates to the identification of biomarkers for gastrointestinal diseases and provides methods utilizing the biomarkers for in drug discovery, monitoring of treatment efficacy, and diagnostics. The invention further provides methods for identifying a therapeutic target to treat ulcerative colitis, colorectal cancer, and Crohn's disease.
Owner:ALFAGENE BIOSCI
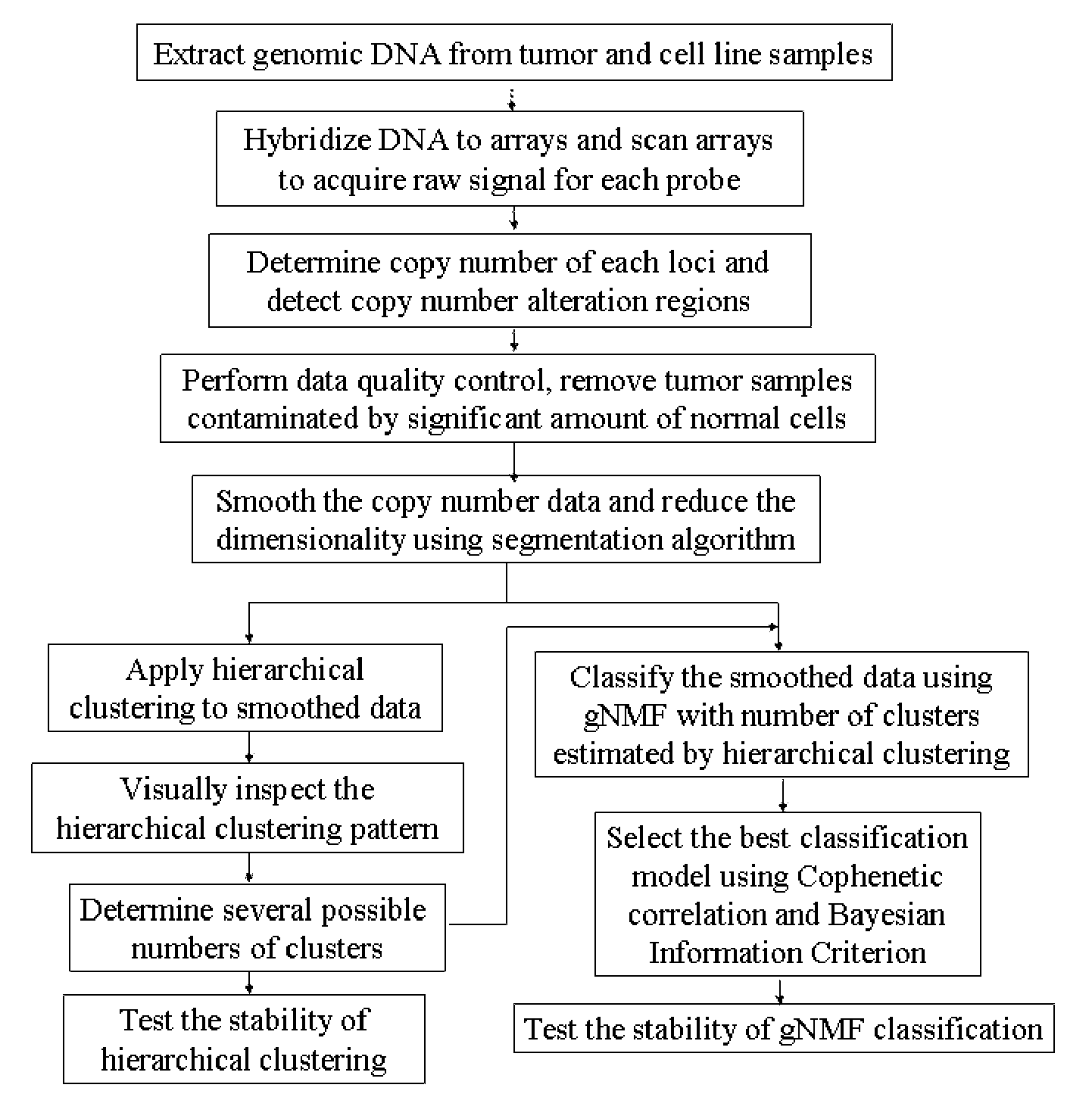
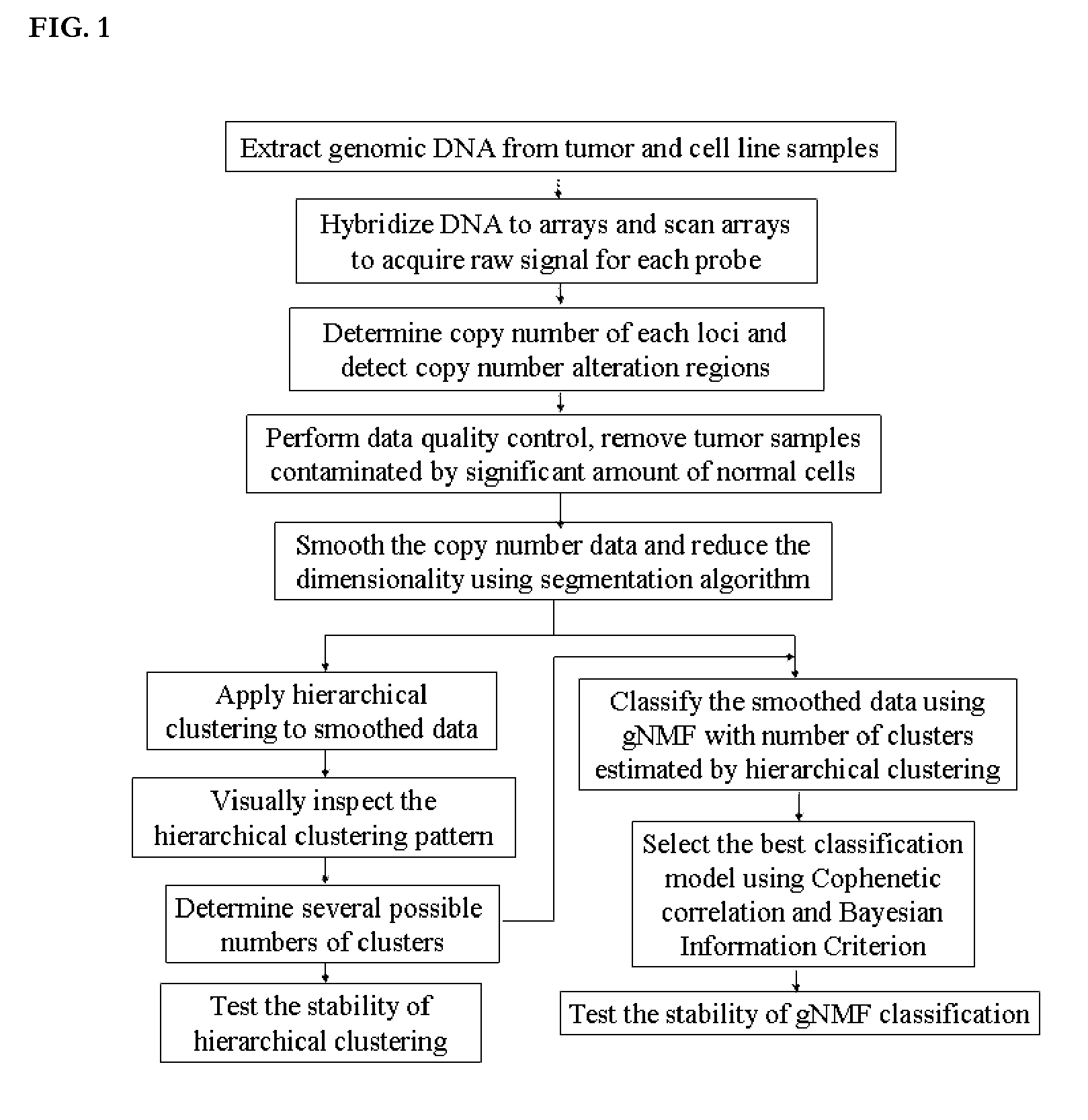
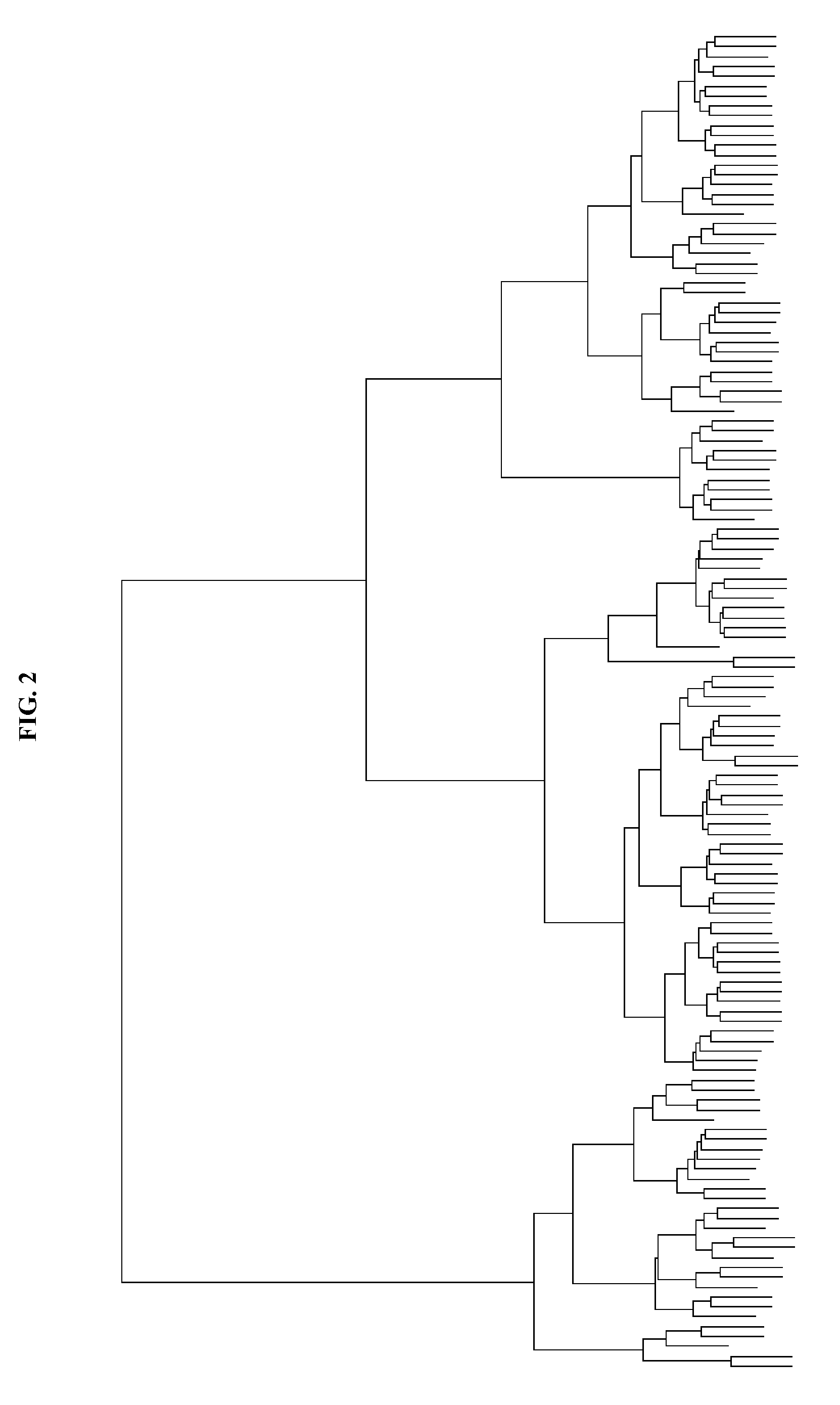
Genomic classification of colorectal cancer based on patterns of gene copy number alterations
The invention is directed to methods and kits that allow for classification of colorectal cancer cells according to genomic profiles, and methods of diagnosing, predicting clinical outcomes, and stratifying patient populations for clinical testing and treatment using the same.
Owner:ABBVIE INC
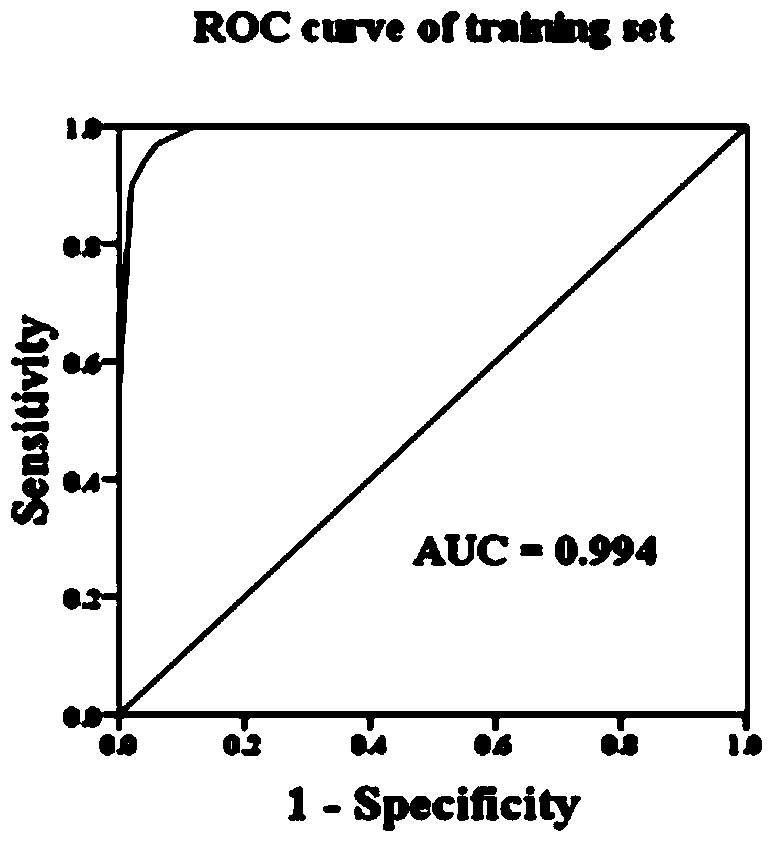
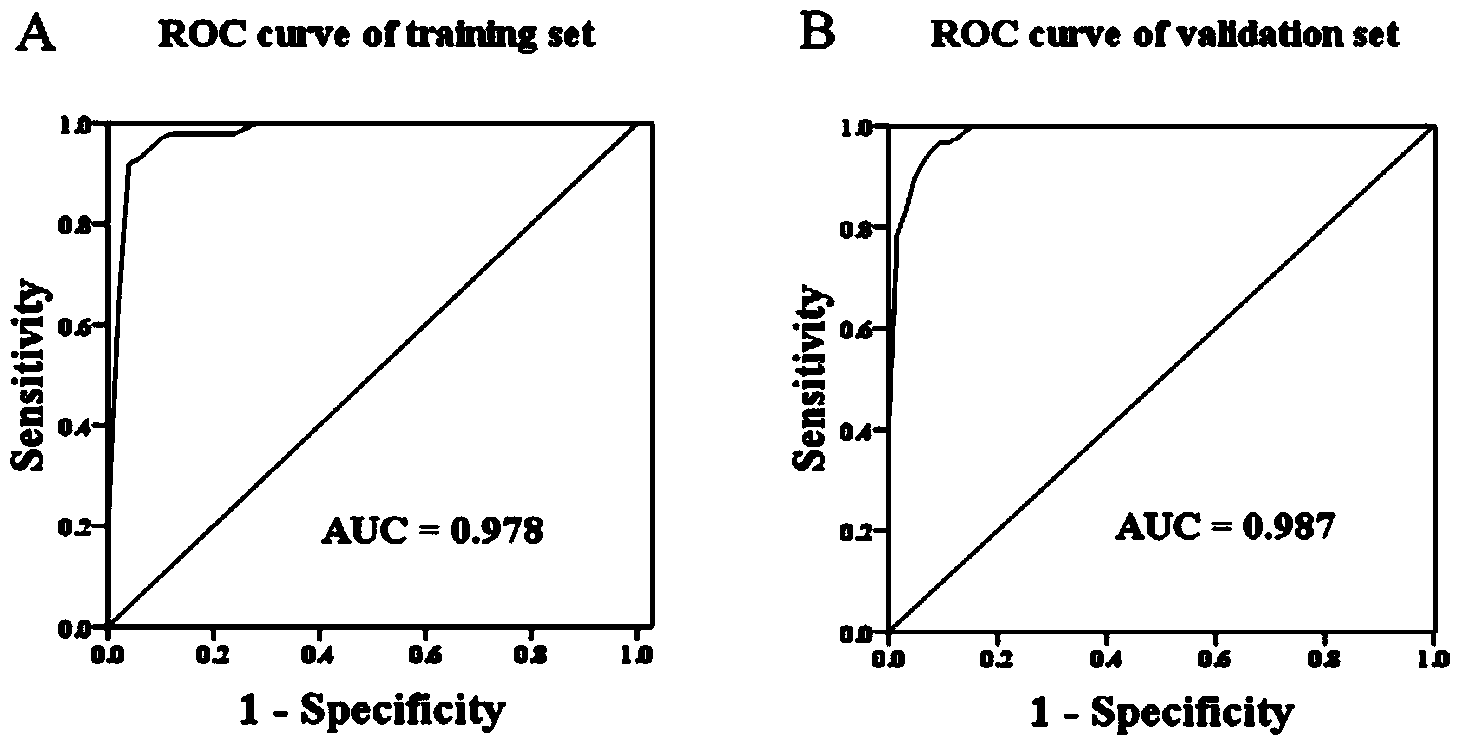
LncRNA biomarkers for diagnosing human lung adenocarcinoma and human colorectal cancer
InactiveCN104131108AIncrease salienceHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationAdenocarcinoma lung cancerTrue positive rate
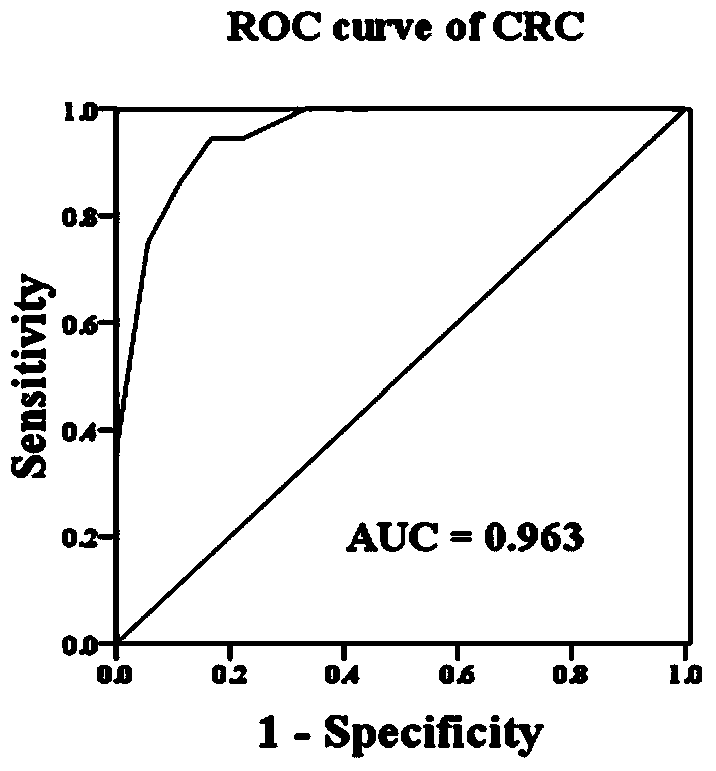
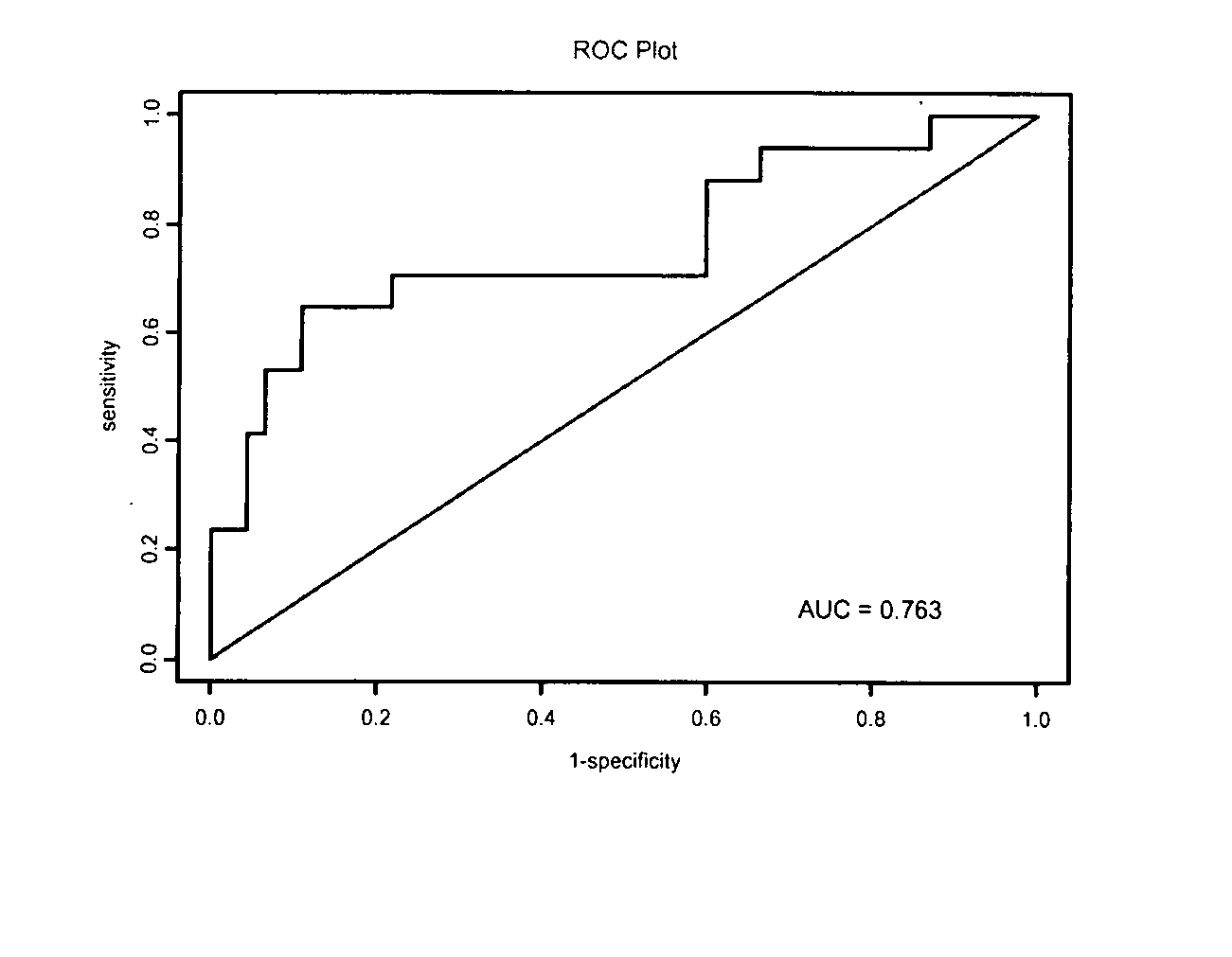
The invention relates to the technical field of biology, and in particular relates to a group of LncRNA biomarkers for diagnosing human lung adenocarcinoma and human colorectal cancer, related kits and application thereof. The group of LncRNA biomarkers for diagnosing human lung adenocarcinoma and / or human colorectal cancer comprises uc001gzl.3, ENST00000434223, uc004bbl.1, ENST00000540136 and NR_034174. The LncRNA biomarkers for diagnosing the human lung adenocarcinoma and / or human colorectal cancer can be used for distinguishing human early lung adenocarcinoma and human colorectal cancer, the AUC of the biomarkers for distinguishing early lung adenocarcinoma from normally paired lung tissues can reach 0.978, and the sensitivity and the specificity are respectively 92% and 98%. The AUC of the biomarkers for diagnosing a colorectal cancer sample can reach 0.963, and the sensitivity and the specificity are respectively 94.4% and 88.9%.
Owner:SHANGHAI INST OF MICROSYSTEM & INFORMATION TECH CHINESE ACAD OF SCI
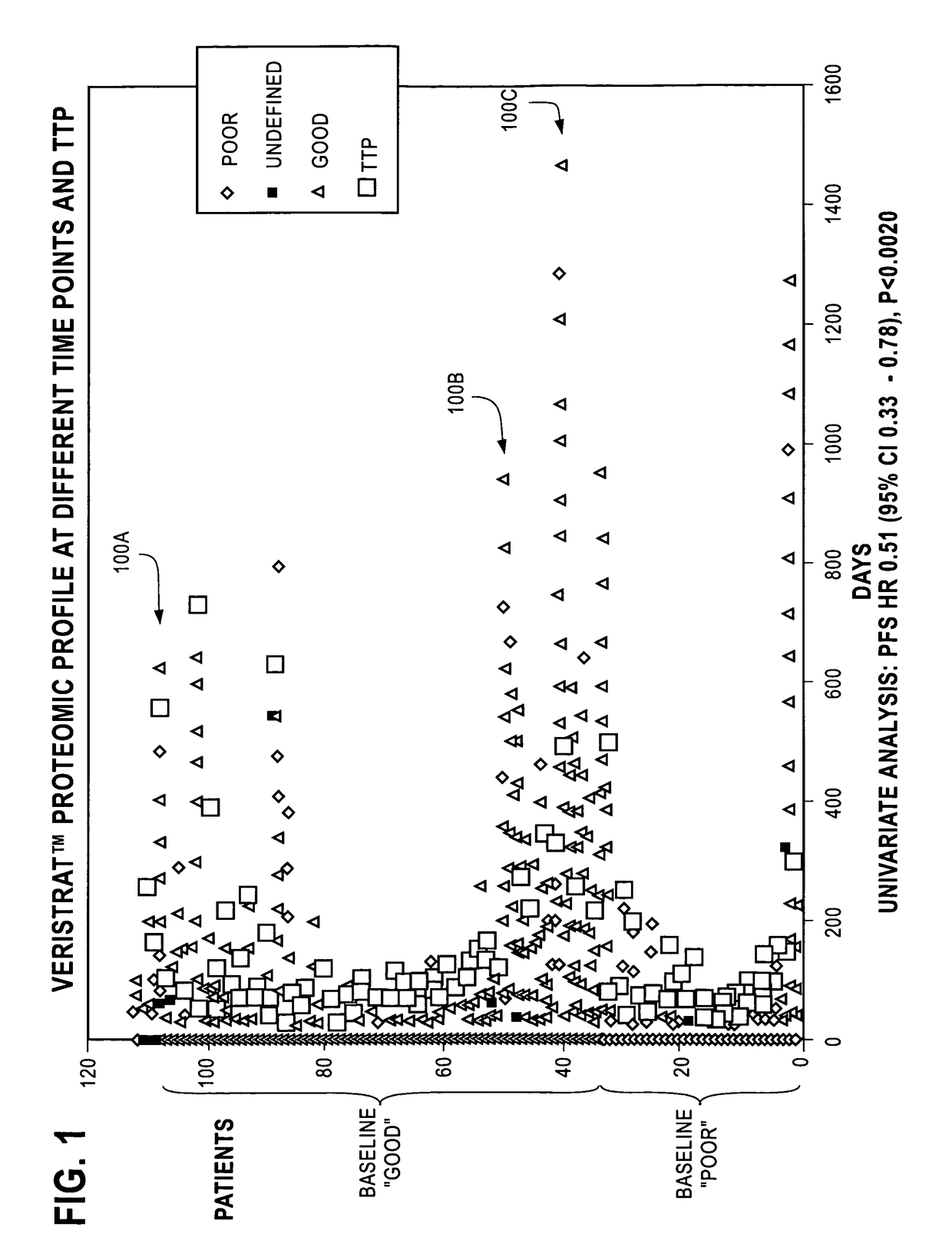
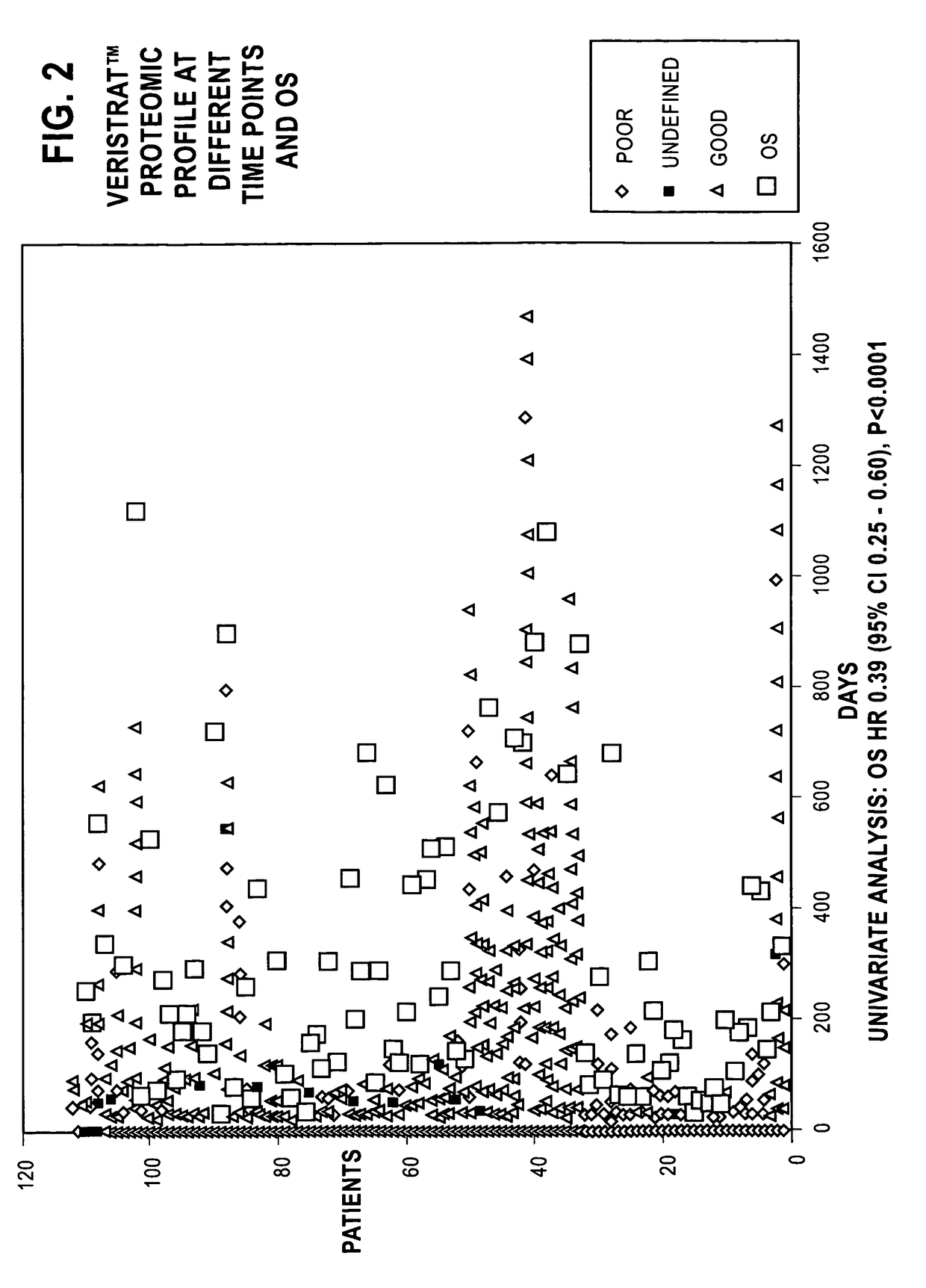
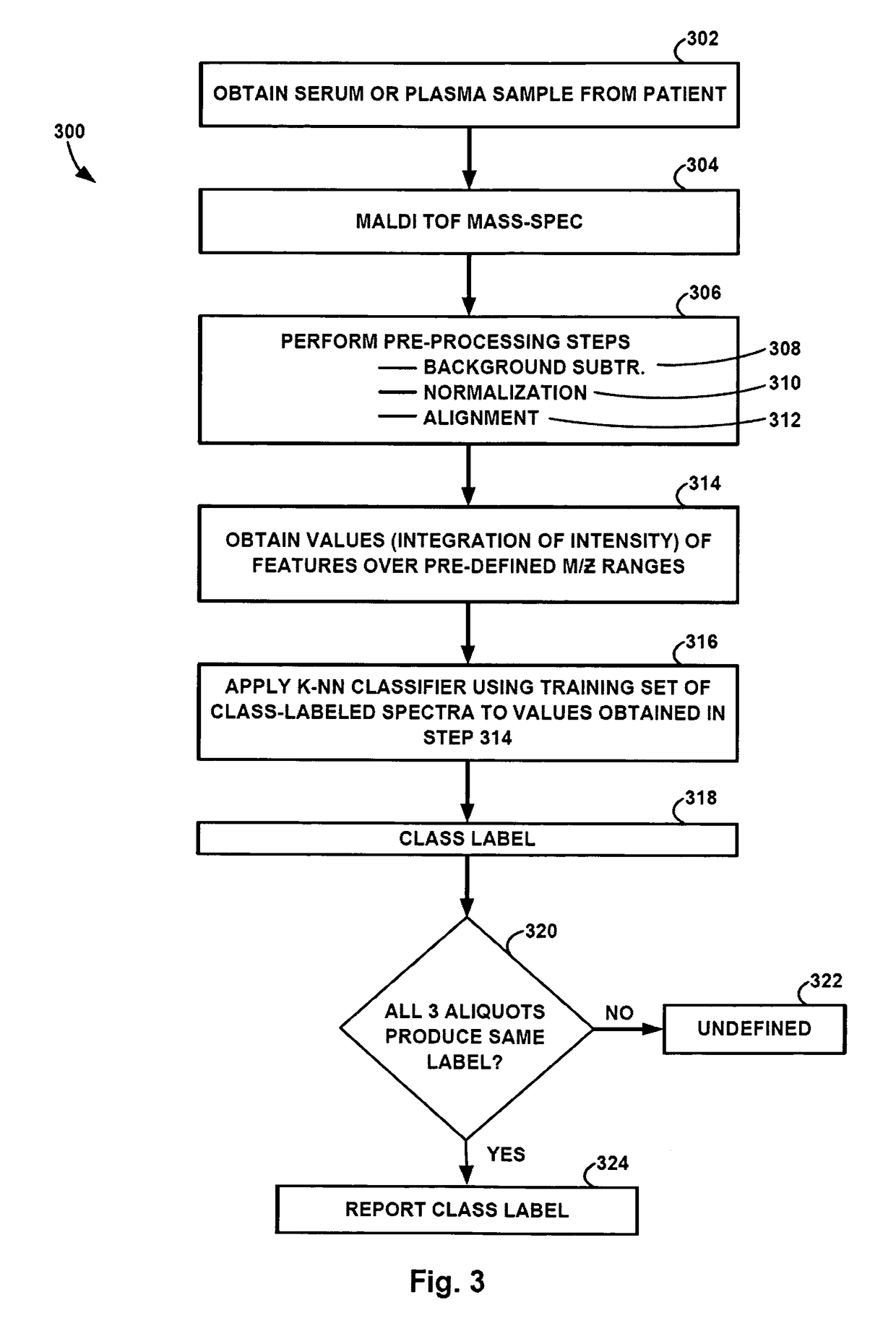
Monitoring treatment of cancer patients with drugs targeting EGFR pathway using mass spectrometry of patient samples
Owner:BIODESIX
Reverse-turn mimetics and method relating thereto
ActiveUS20060084655A1Efficient implementationPromoting neurite outgrowthBiocideNervous disorderUlcerative colitisNodular sclerosis
Conformationally constrained compounds that mimic the secondary structure of reverse-turn regions of biologically active peptides and proteins are disclosed. Such reverse-turn mimetic structures have utility over a wide range of fields, including use as diagnostic and therapeutic agents. Libraries containing the reverse-turn mimetic structures of this invention are also disclosed as well as methods for screening the same to identify biologically active members. The invention also relates to the use of such compounds for inhibiting or treating disorders modulated by Wnt-signaling pathway, such as cancer, especially colorectal cancer, restenosis associated with angioplasty, polycystic kidney disease, aberrant angiogenesis disease, rheumatoid arthritis disease, tuberous sclerosis complex, Alzheimer's disease, excess hair growth or loss, or ulcerative colitis.
Owner:JW PHARMA CORP
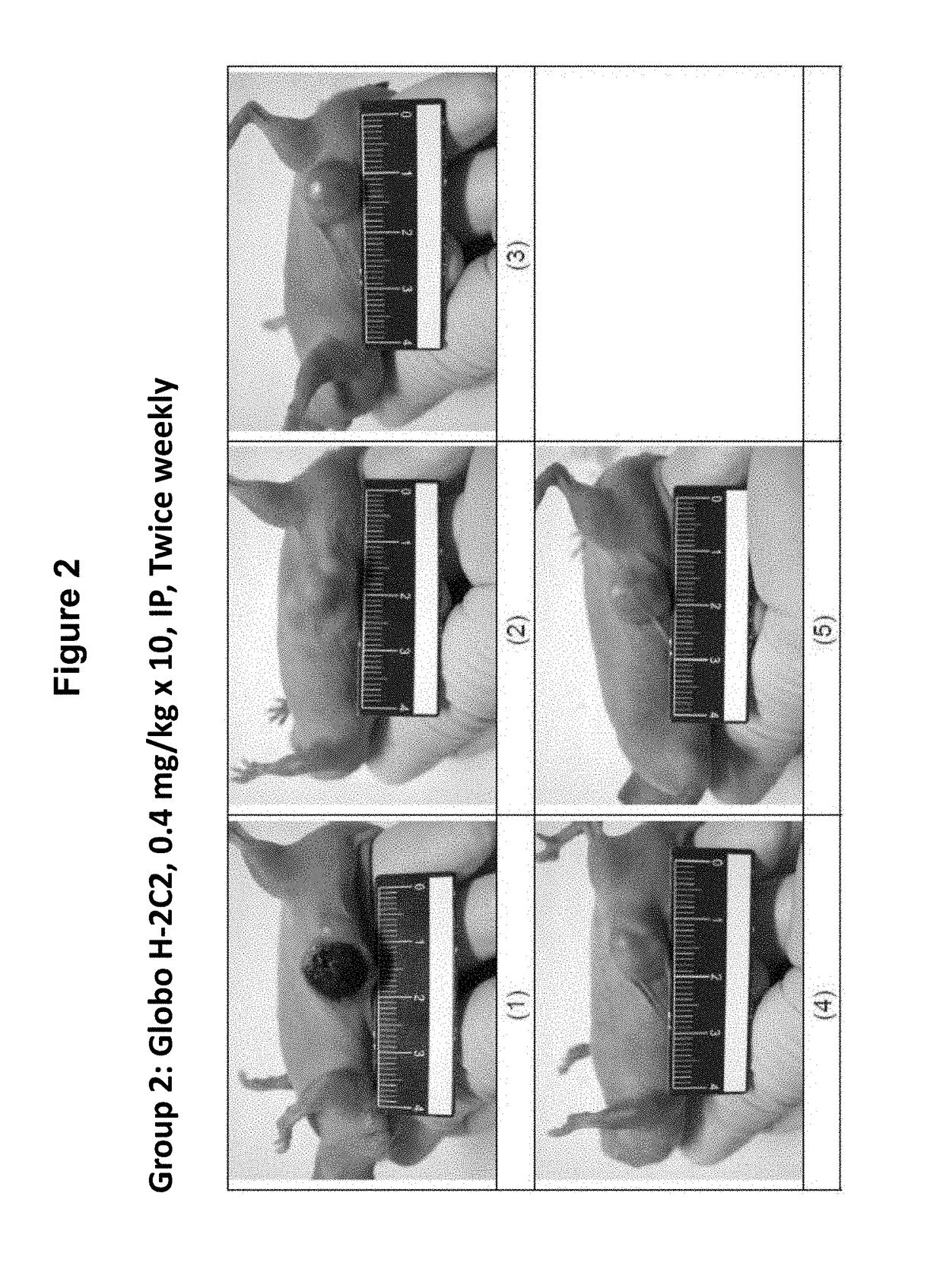
Antibodies, pharmaceutical compositions and methods
ActiveUS20170283488A1Polypeptide with localisation/targeting motifAntibody mimetics/scaffoldsURINARY BLADDER CARCINOMASquamous Carcinomas
Pharmaceutical composition comprising antibodies or antigen binding fragments thereof that bind to stage-specific embryonic antigen 4 (SSEA-4) are disclosed herein, as well as methods of use thereof. Methods of use include, without limitation, cancer therapies and diagnostics. The antibodies of the disclosure can bind to certain cancer cell surfaces. Exemplary targets of the antibodies disclosed herein can include carcinomas, such as breast cancer, lung cancer, esophageal cancer, rectal cancer, biliary cancer, liver cancer, buccal cancer, gastric cancer, colon cancer, nasopharyngeal cancer, kidney cancer, prostate cancer, ovarian cancer, cervical cancer, endometrial cancer, pancreatic cancer, testicular cancer, bladder cancer, head and neck cancer, oral cancer, neuroendocrine cancer, adrenal cancer, thyroid cancer, bone cancer, skin cancer, basal cell carcinoma, squamous cell carcinoma, melanoma, and / or brain tumor.
Owner:OBI PHARMA INC
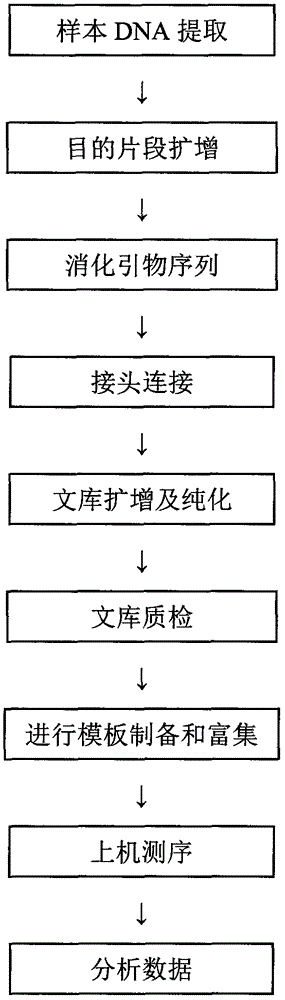
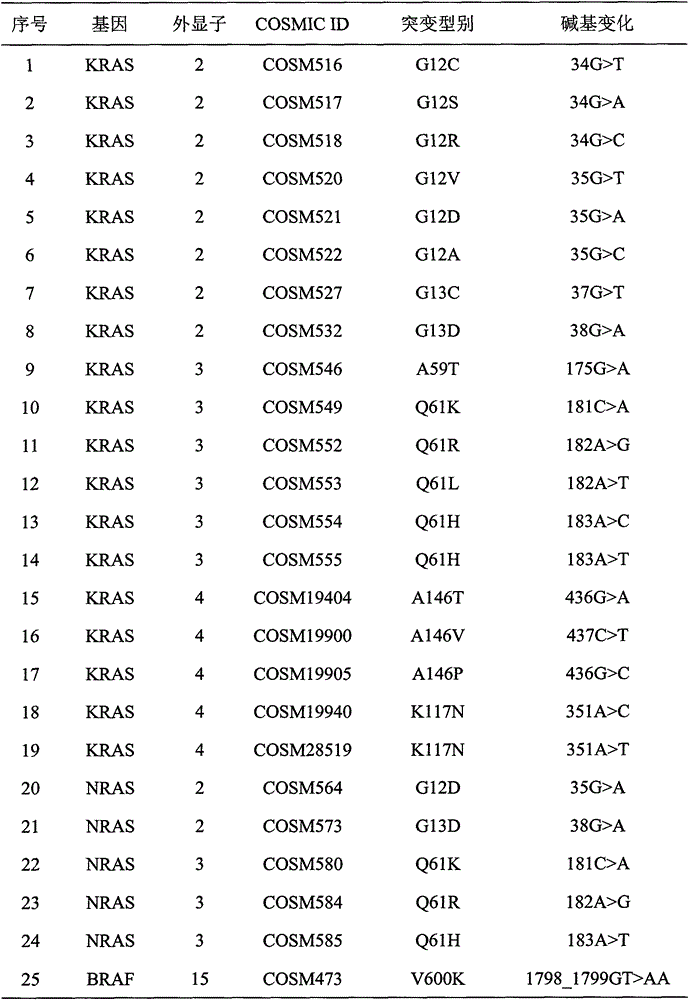
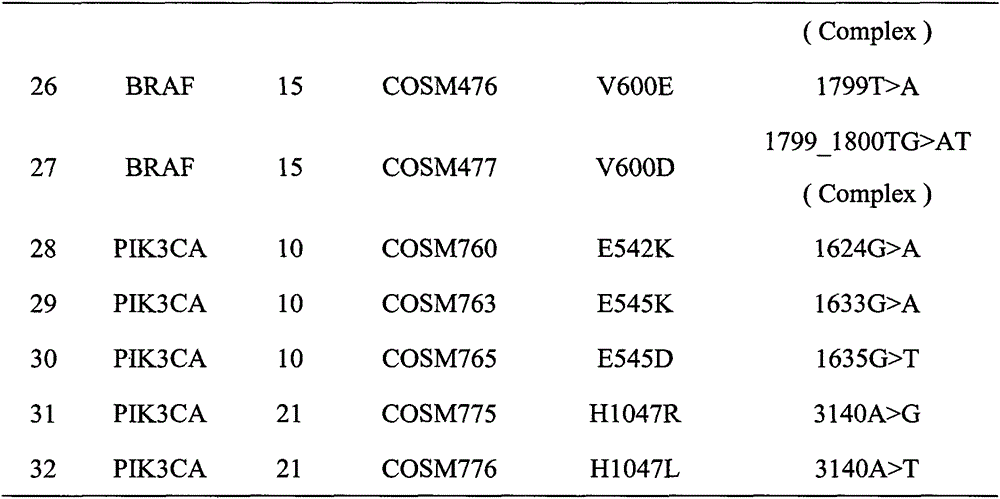
Method and kit for detecting colorectal cancer KRAS/NRAS/BRAF/PIK3CA genes
ActiveCN106755445AImprove efficiencyReduced sensitivity and accuracyMicrobiological testing/measurementDigestionBiotechnology
The invention relates to the field of biotechnology, in particular to a method and kit for detecting colorectal cancer KRAS / NRAS / BRAF / PIK3CA genes based on a high-throughput sequencing platform. The method comprises the steps of extraction of sample DNA, amplification of a target fragment, digestion of a primer sequence, connection of joints, amplification and purification of a library, quality testing of the library, template preparation and enrichment, online sequencing and data analysis. By the adoption of the method and kit for detecting colorectal cancer KRAS / NRAS / BRAF / PIK3CA genes, detection efficiency, accuracy and sensitivity are greatly improved, detection cost is reduced, and a more definite basis can be provided for individualized treatment of colorectal cancer patients.
Owner:GUANGZHOU DARUI BIOTECH
Colorectal cancer diagnostic biomarker and application thereof
ActiveCN107447033AAccurate judgmentQuick judgmentNucleotide librariesMicrobiological testing/measurement5 year survival rateMortality rate
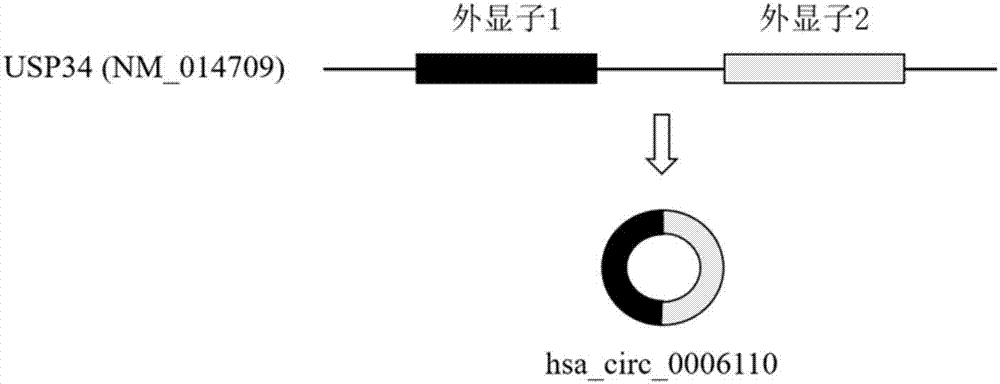
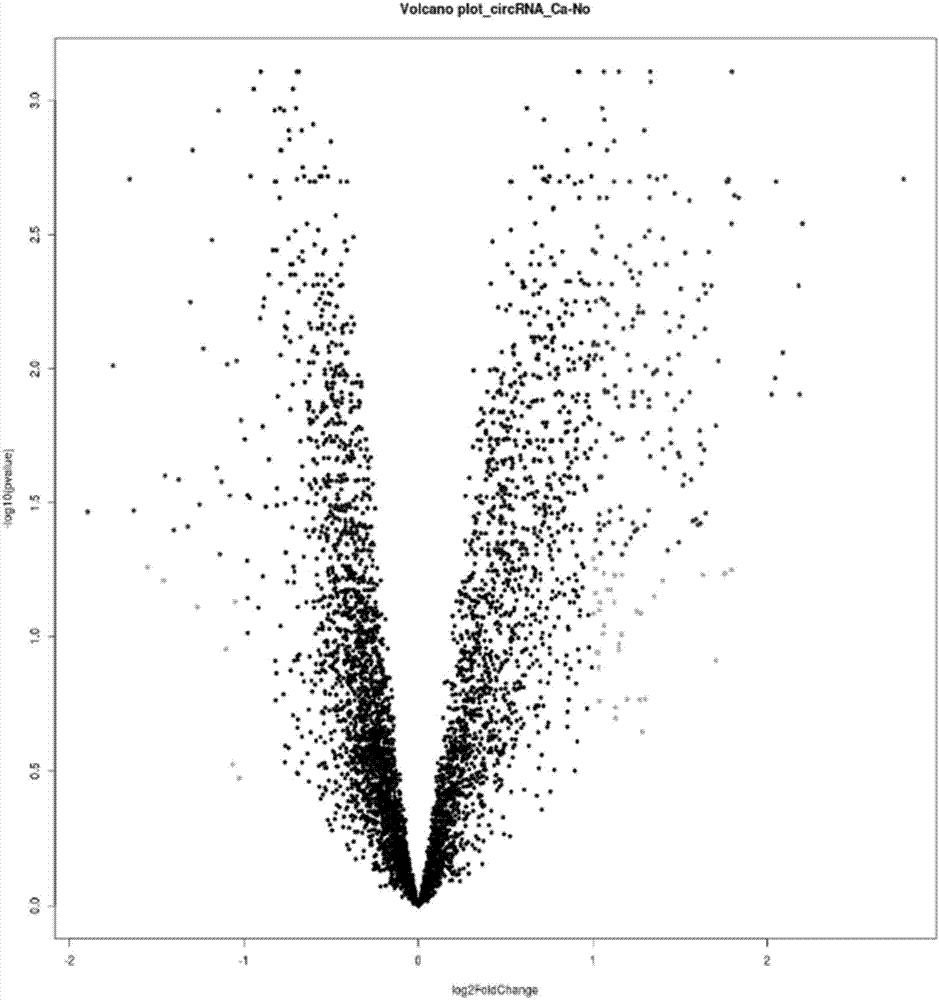
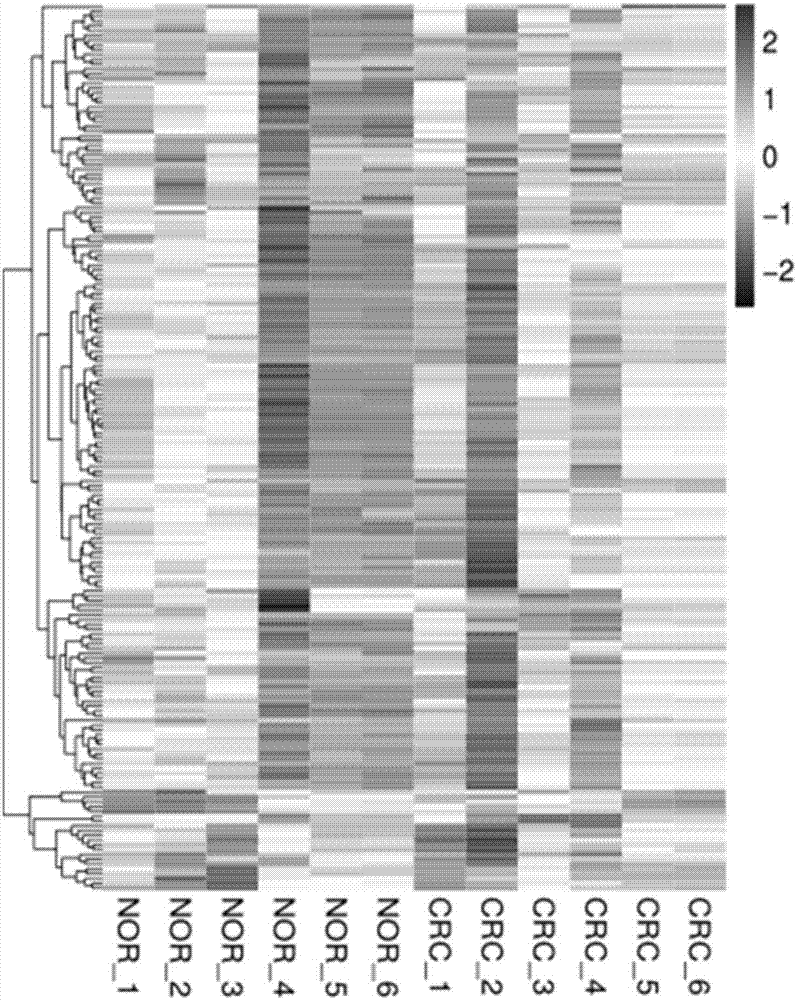
The invention relates to a colorectal cancer diagnostic biomarker and application thereof. A nucleotide sequence is shown as SEQ ID NO. 1. The colorectal cancer diagnostic biomarker provided by the invention has the beneficial effects that hsa_circ_0006110 gene expression is discovered to be closely related to a colorectal cancer for the first time; through detecting the expression of hsa_circ_0006110 in a subject colorectal tissue, whether a subject suffers from the colorectal cancer or the risk of the colorectal cancer exists or not can be more accurately and quickly judged, so that a prevention or treatment scheme is provided for a clinician. A target gene as a drug for treating the colorectal cancer provides a new treatment targets and treatment approach for colorectal cancer treatment; compared with a traditional detection means, the diagnostic biomarker is more timely and more specific to diagnose, so that the five-year survival rate of a colorectal cancer patient is improved, and the death rate is reduced, therefore, the application prospect is wide.
Owner:SOUTHEAST UNIV
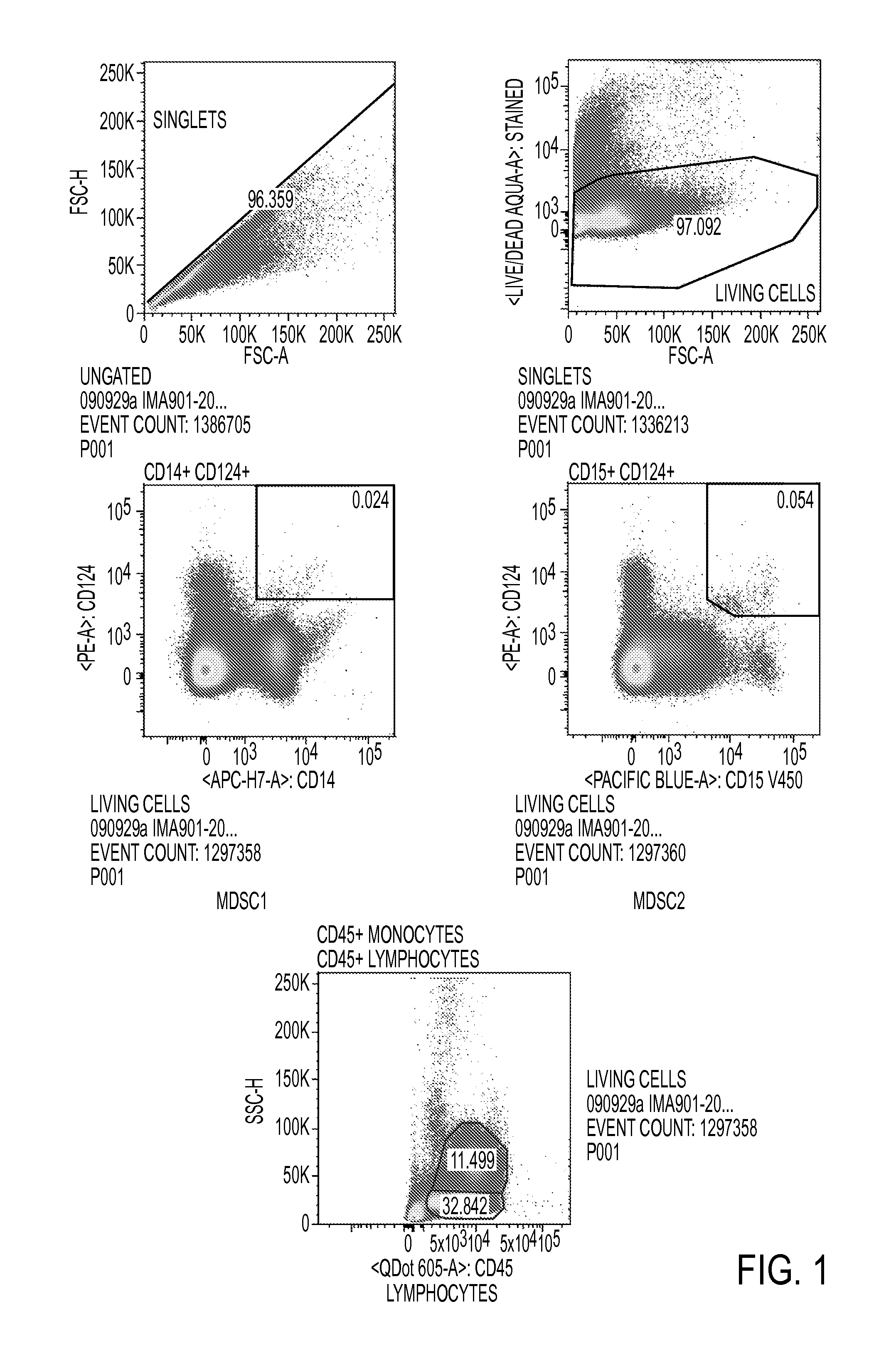
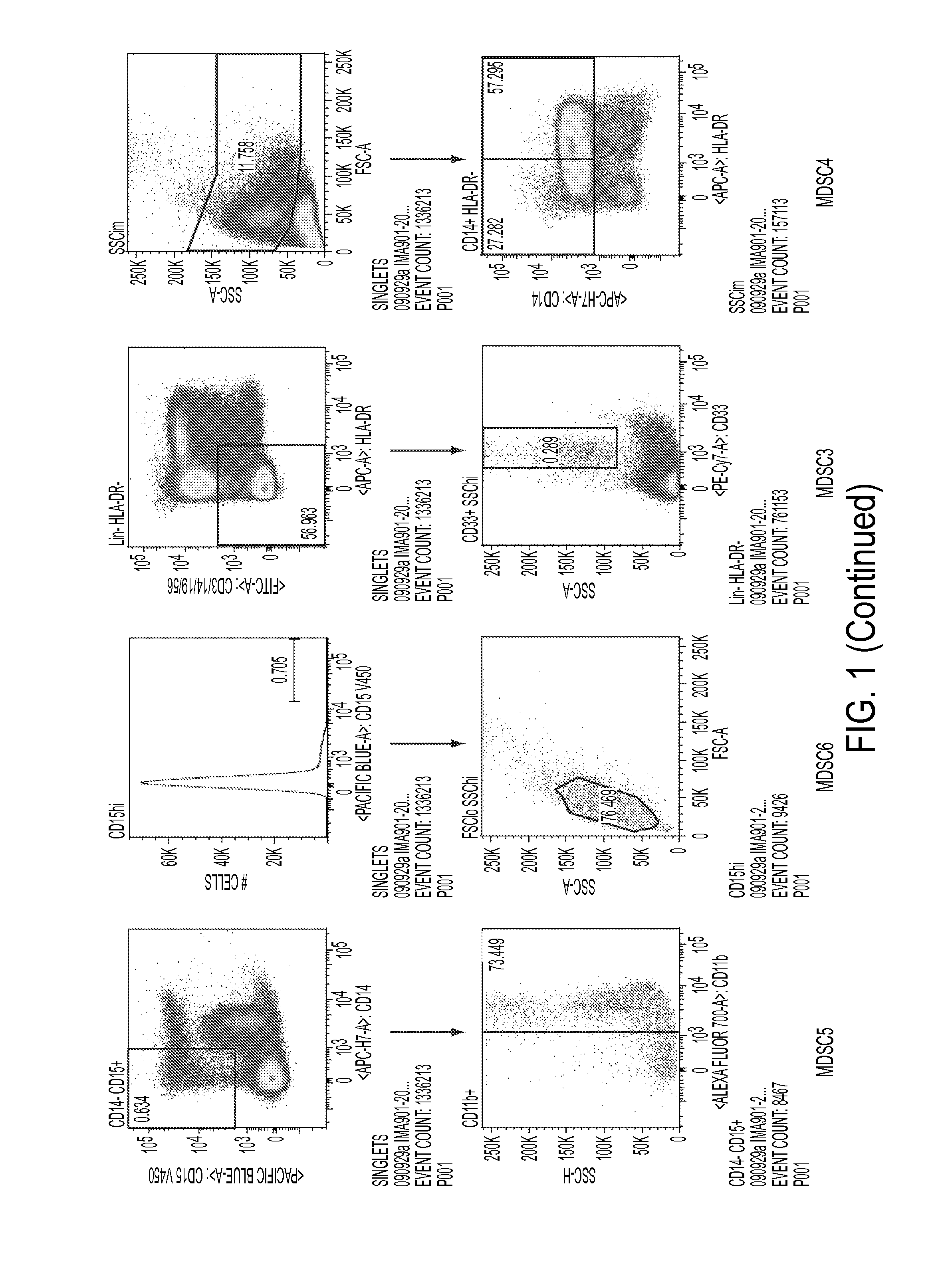
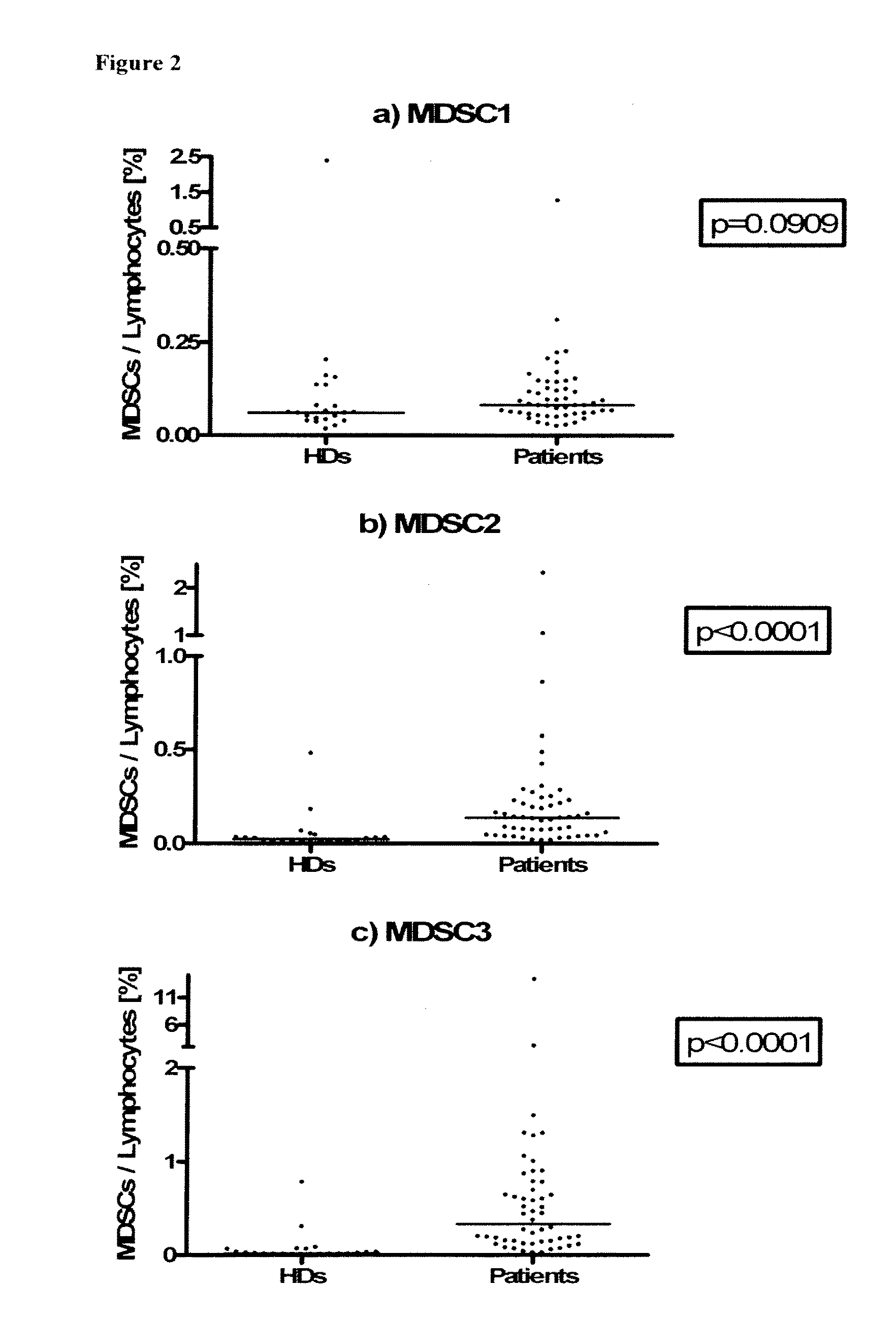
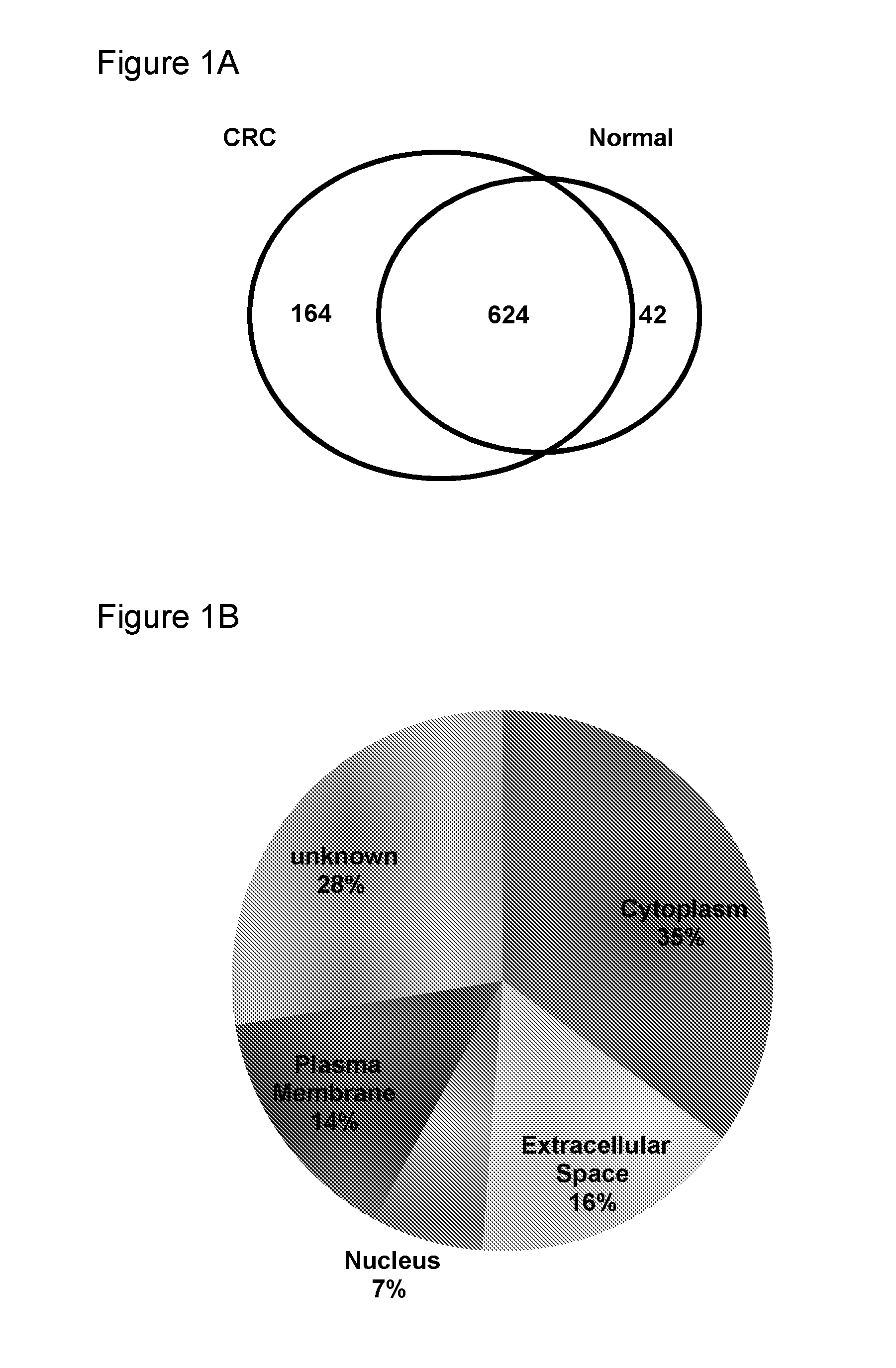
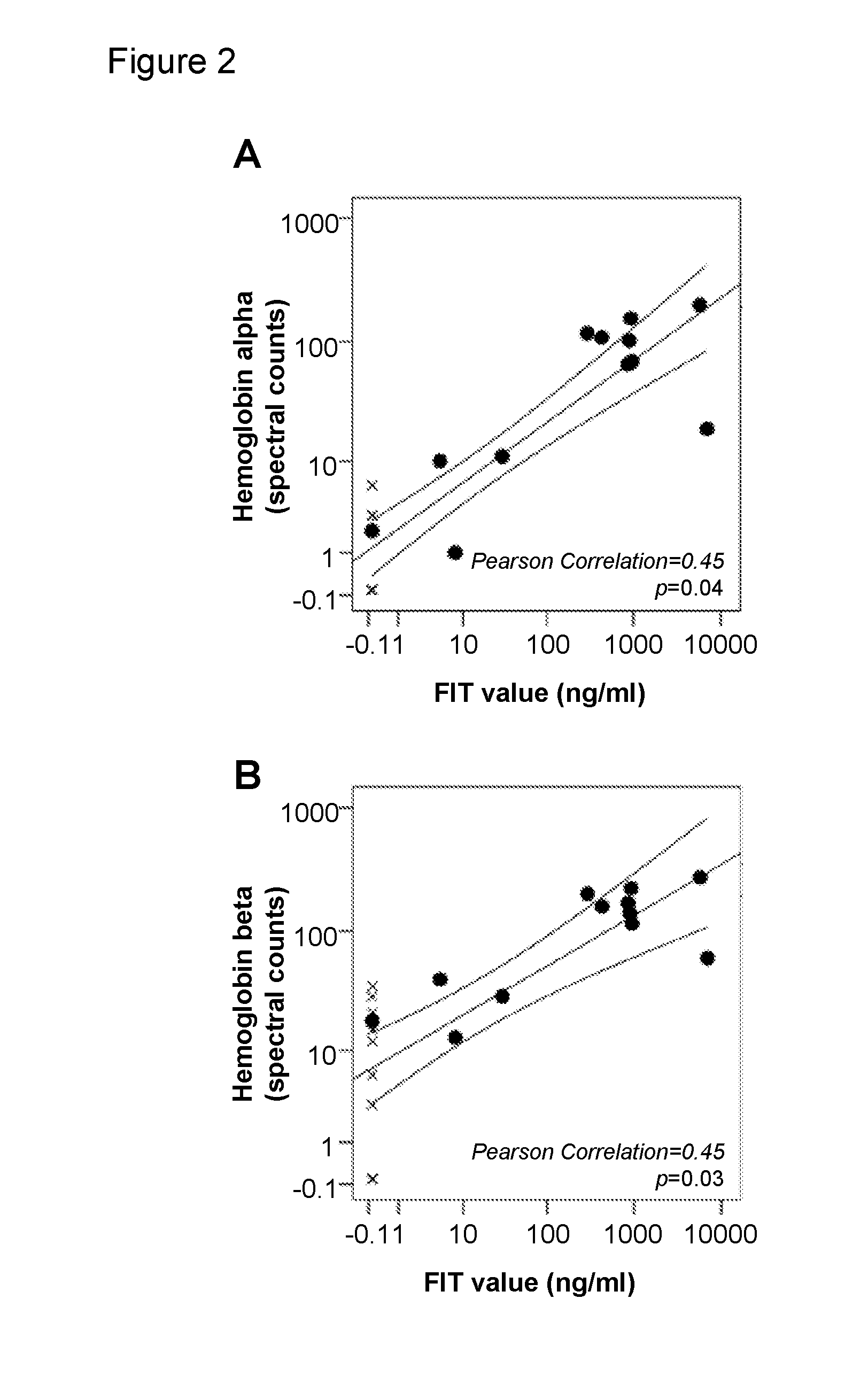
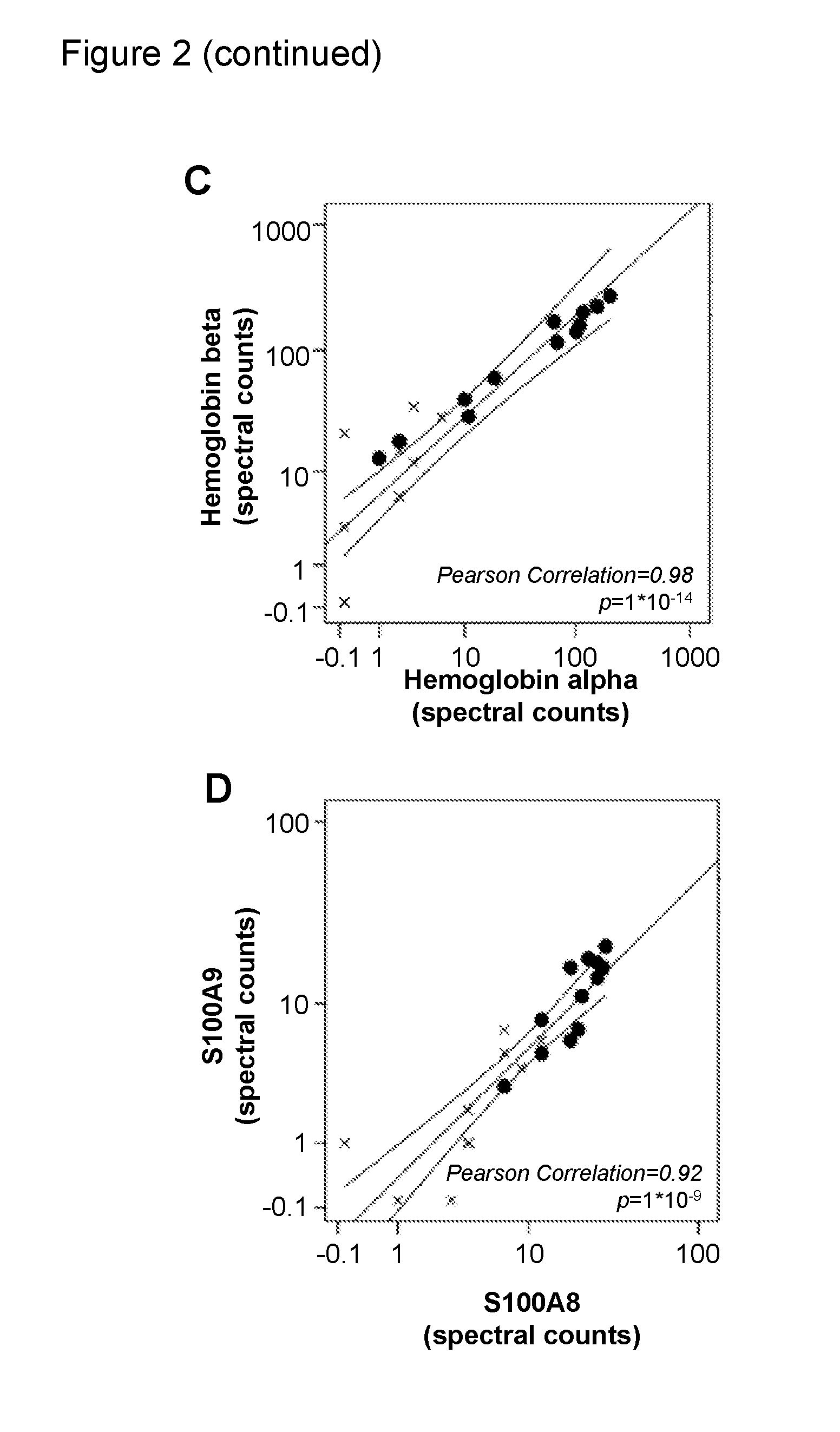
Use of myeloid cell biomarkers for the diagnosis of cancer
ActiveUS20120070461A1Diagnosing and prognosing cancerImprove the level ofBiological testingImmunological disordersCancers diagnosisTherapeutic effect
The present invention relates to the use of myeloid cell biomarkers for the differential diagnosis, prognosis, and monitoring of renal cell carcinoma (RCC) or colorectal cancer (CRC). The present invention furthermore relates to monitoring the effect of a treatment against renal cell carcinoma (RCC) or colorectal cancer (CRC), and establishing a prognosis of the outcome of the treatment of renal cell carcinoma (RCC) or colorectal cancer (CRC). The present invention furthermore relates to panels of cellular biomarkers for use in the above methods, in particular multicolor panels for measuring said biomarkers.
Owner:IMMATICS BIOTECHNOLOGIES GMBH
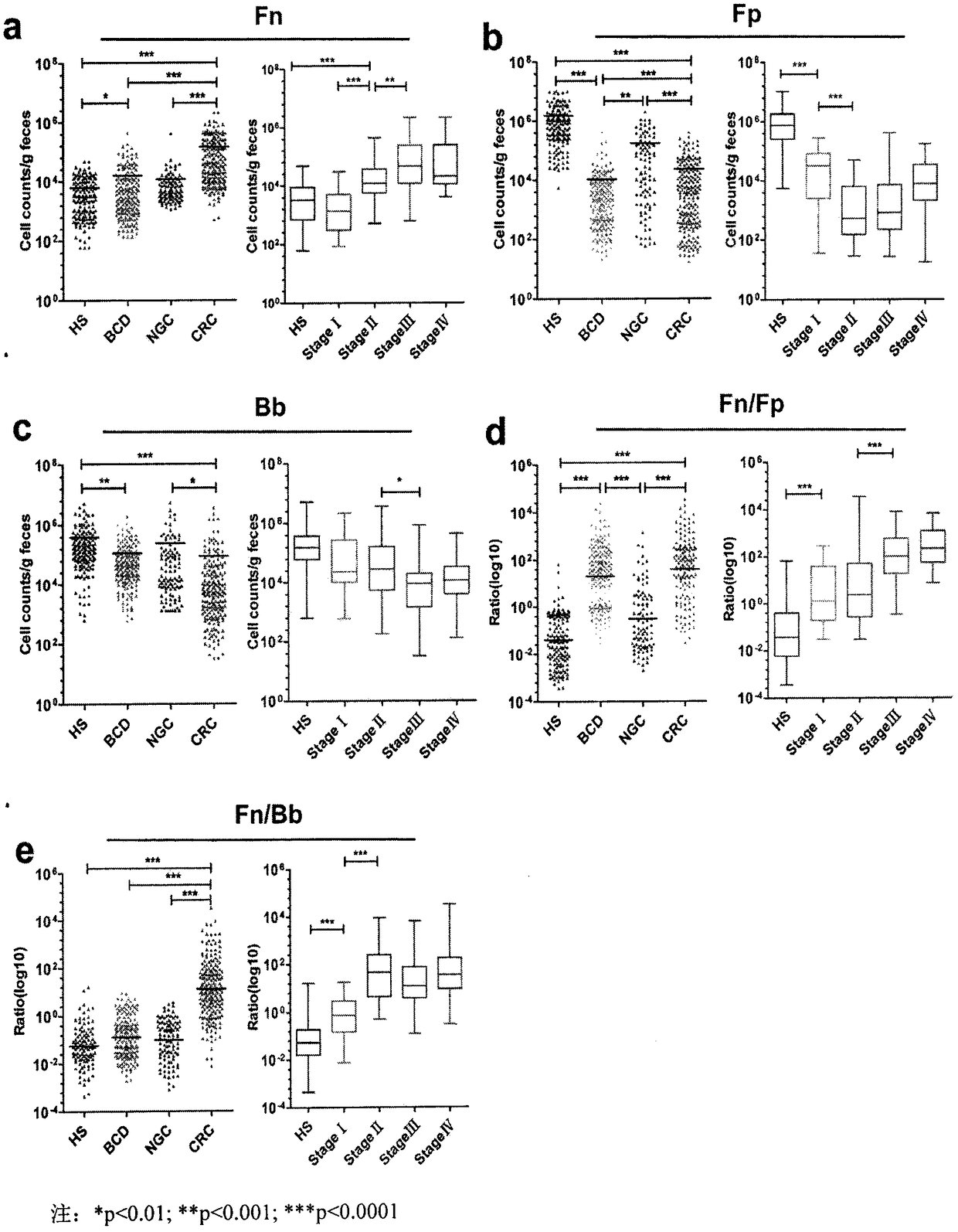
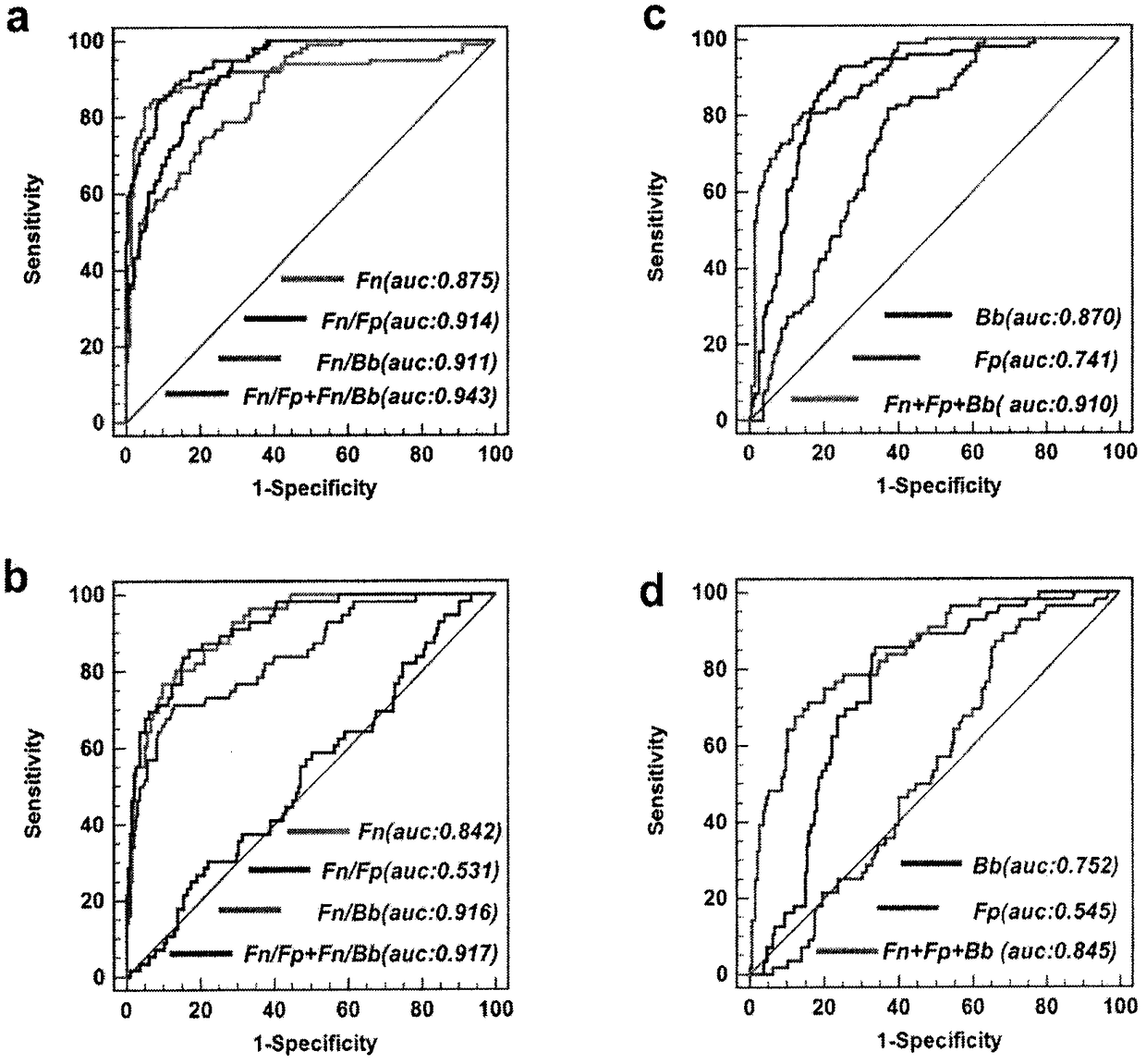
Method for evaluating stable state of flora in excrement sample and application of method in colorectal cancer screening
PendingCN108690864ALow costGood Gut Health ScreeningMicrobiological testing/measurementMicroorganism based processesClostridium leptumFeces
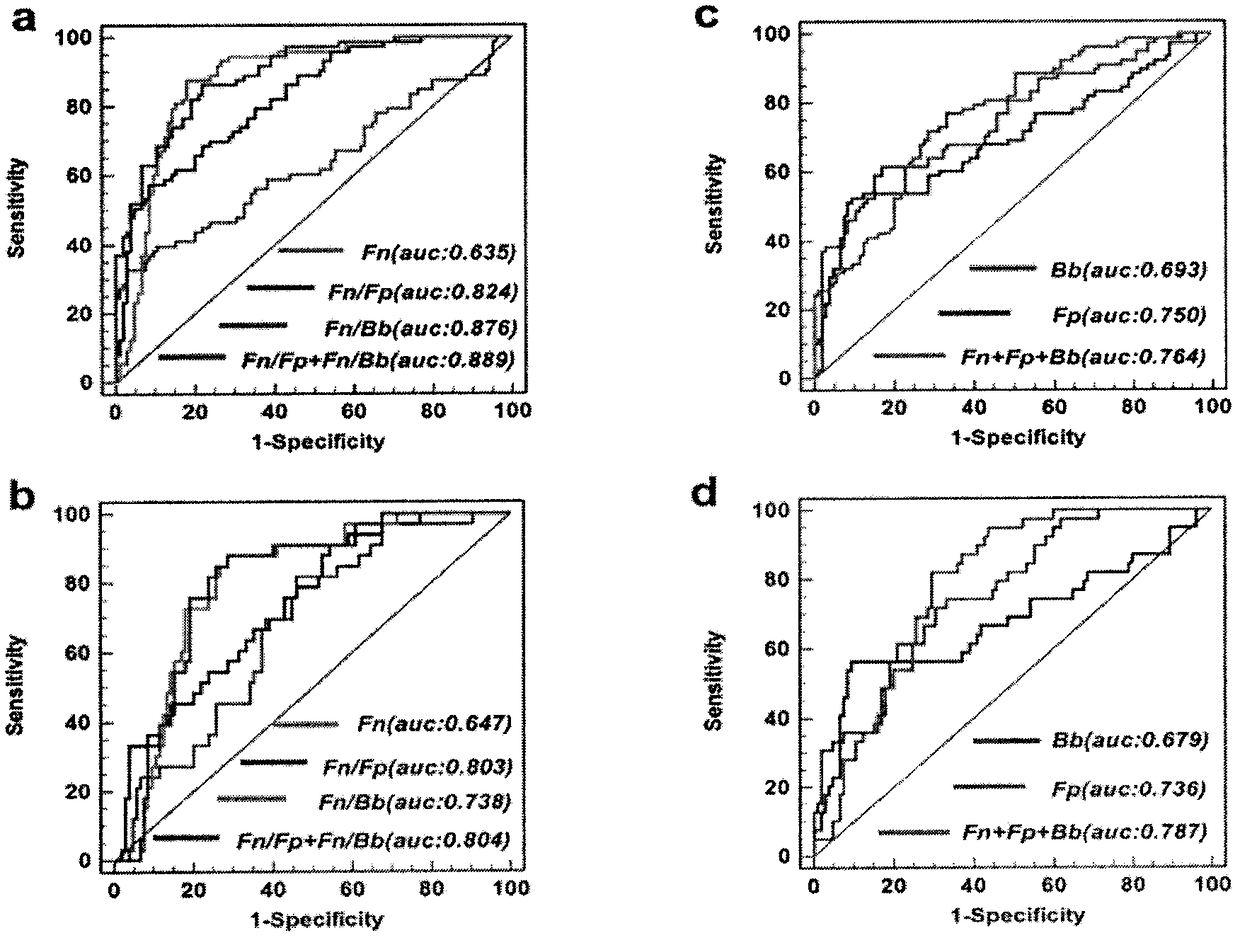
The invention relates to a method for calculating a flora balanced relation index in an individual excrement sample, and an application of the method in screening, diagnosis or auxiliary diagnosis incolorectal cancer (CRC). By extracting bacteria in excrement during DNA sequencing, the types and quantity characteristics of the bacteria can be obtained, and a CRC diagnosis by taking quantity ratiocharacteristic of a plurality of bacteria as a base is carried out. Compared with the methods used in clinical diagnosis or noninvasive screening of CRC with an applied patent, the method is completely noninvasive, and can realize accurate diagnosis of CRC. The analysis result displays that a ratio of fusobacterium nucleatum Fn to bifidobacteria Bb quantity (Fn / Bb) has high susceptibility and specificity on CRC screening, which can respectively reach 84.6% and 92.3% (AUC=0.911). the ratio of fusobacterium nucleatum Fn to clostridium leptum Fp (Fn / Fp) quantity is combined to increase the diagnosis value on CRC, and the Area Under Curve (AUC) of a subject work characteristic curve can reach 0.943. In addition, combination of Fn / Bb and Fn / Fp quantity ratio for screening I-stage CRC has 60% of specificity and 90% of sensitivity.
Owner:SUN YAT SEN UNIV
Long noncoding RNA (LncRNA), and primer pair and kit for detecting expression level of long noncoding RNA in cells and tissues
ActiveCN104877998AOriginalSimple and fast operationMicrobiological testing/measurementDNA/RNA fragmentationCarcinomaBiology
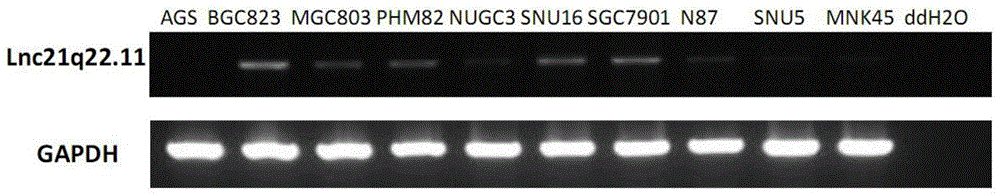
The invention provides a new long noncoding RNA (LncRNA) Lnc21q22.11 sequence, and a primer pair and kit for detecting expression level of the long noncoding RNA in cells and tissues. The full length of the LncRNA Lnc21q22.11 is identified to be 1202bp for the first time, the specific primers are utilized for the first time to detect the expression level of the LncRNA Lnc21q22.11 in stomach cancer cells, 35 pairs of stomach cancer line and para-carcinoma tissues, colorectal cancer cell lines and 10 pairs of colorectal cancer and para-carcinoma tissues, and thus, the kit is creative. The detection of the expression of the LncRNA Lnc21q22.11 in the stomach cancer tissues by using the kit and specific primers can be used as an effective means for stomach cancer and colorectal cancer diagnosis, curative effect observation, prognosis judgment, focus residue and relapse detection and the like. The kit is simple to operate, has the advantages of high stability and high sensitivity, and has far-reaching clinical meanings and popularization performance.
Owner:GENERAL HOSPITAL OF PLA
Chemoradiotherapy with TS-1/camptothecins
InactiveUS20070036717A1Improve the level ofLess side effectsBiocideIn-vivo radioactive preparationsAbnormal tissue growthSide effect
An antitumor agent for chemoradiotherapy of rectal cancer comprising a combination of TS-1 (a combination drug of tegafur, gimeracil, and oteracil potassium at a 1:0.4:1 molar ratio) and CPT-11 (irinotecan hydrochloride). The antitumor agent of the invention can achieve marked tumor volume reduction without causing major side effects, especially by coadministering it with preoperative radiation therapy. The volume of even large tumors that are refractory to surgical resection can be reduced by coadministering the antitumor agent of the invention with preoperative radiation therapy, making the subsequent surgical resection of the tumor easier.
Owner:THE KITASATO INST
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com




![Imidazo[4, 5-B]pyridin-2-one and oxazolo[4, 5-B] pyridin-2-one compounds and analogs thereof as cancer therapeutic compounds Imidazo[4, 5-B]pyridin-2-one and oxazolo[4, 5-B] pyridin-2-one compounds and analogs thereof as cancer therapeutic compounds](https://images-eureka.patsnap.com/patent_img/56ad8e42-9fd9-4d02-ba70-fb173ab812ce/US07951819-20110531-C00001.png)
![Imidazo[4, 5-B]pyridin-2-one and oxazolo[4, 5-B] pyridin-2-one compounds and analogs thereof as cancer therapeutic compounds Imidazo[4, 5-B]pyridin-2-one and oxazolo[4, 5-B] pyridin-2-one compounds and analogs thereof as cancer therapeutic compounds](https://images-eureka.patsnap.com/patent_img/56ad8e42-9fd9-4d02-ba70-fb173ab812ce/US07951819-20110531-C00002.png)
![Imidazo[4, 5-B]pyridin-2-one and oxazolo[4, 5-B] pyridin-2-one compounds and analogs thereof as cancer therapeutic compounds Imidazo[4, 5-B]pyridin-2-one and oxazolo[4, 5-B] pyridin-2-one compounds and analogs thereof as cancer therapeutic compounds](https://images-eureka.patsnap.com/patent_img/56ad8e42-9fd9-4d02-ba70-fb173ab812ce/US07951819-20110531-C00003.png)