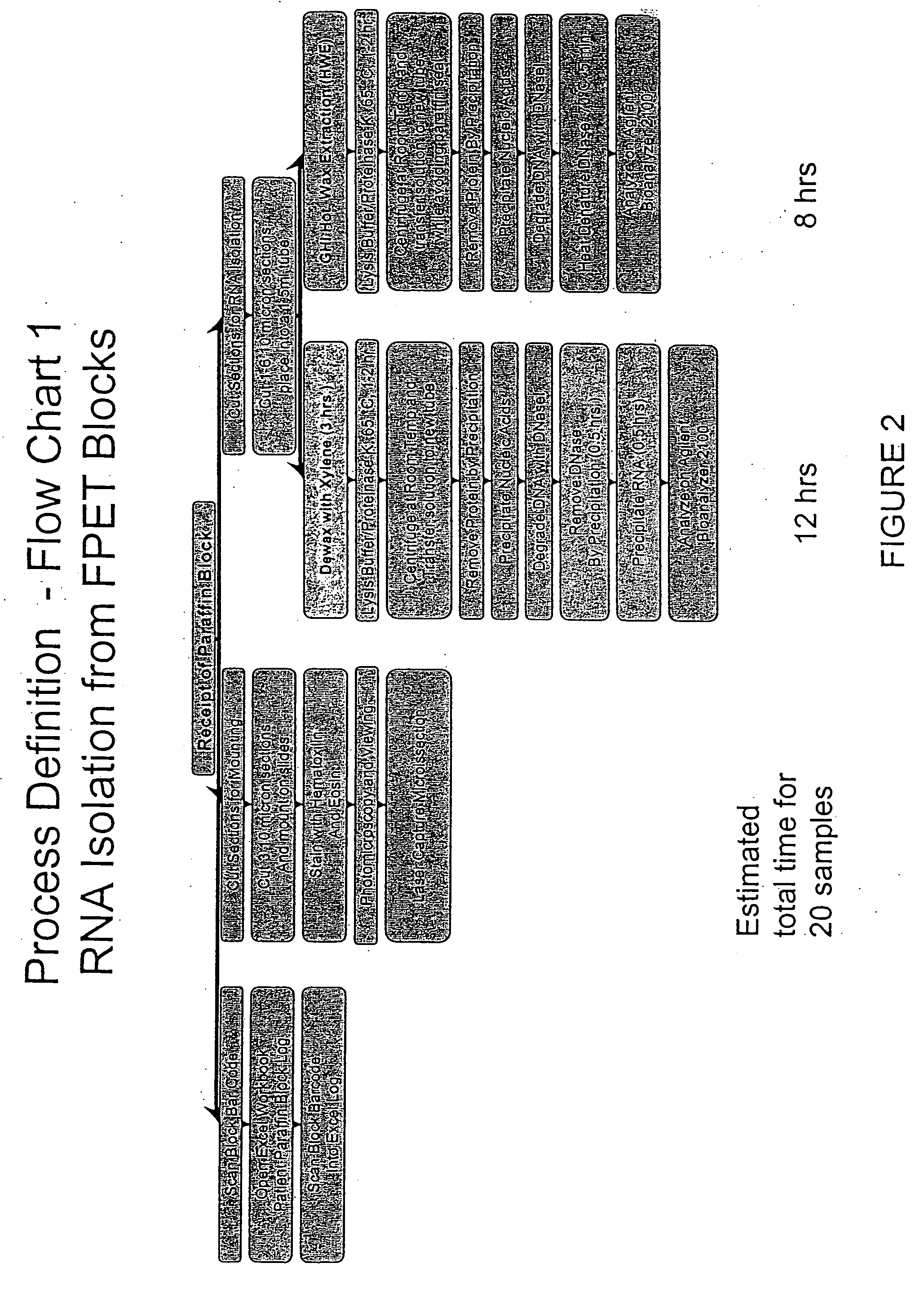
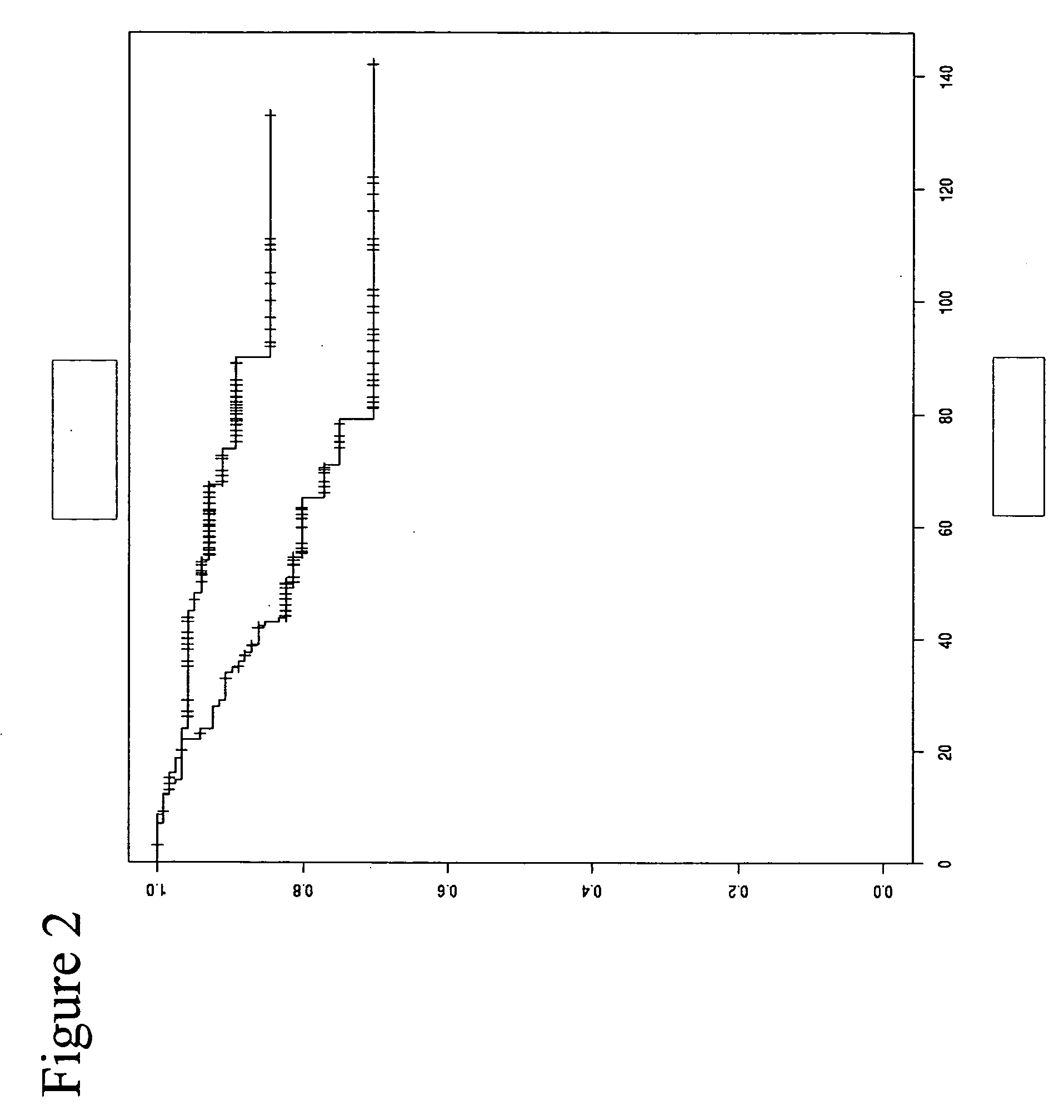
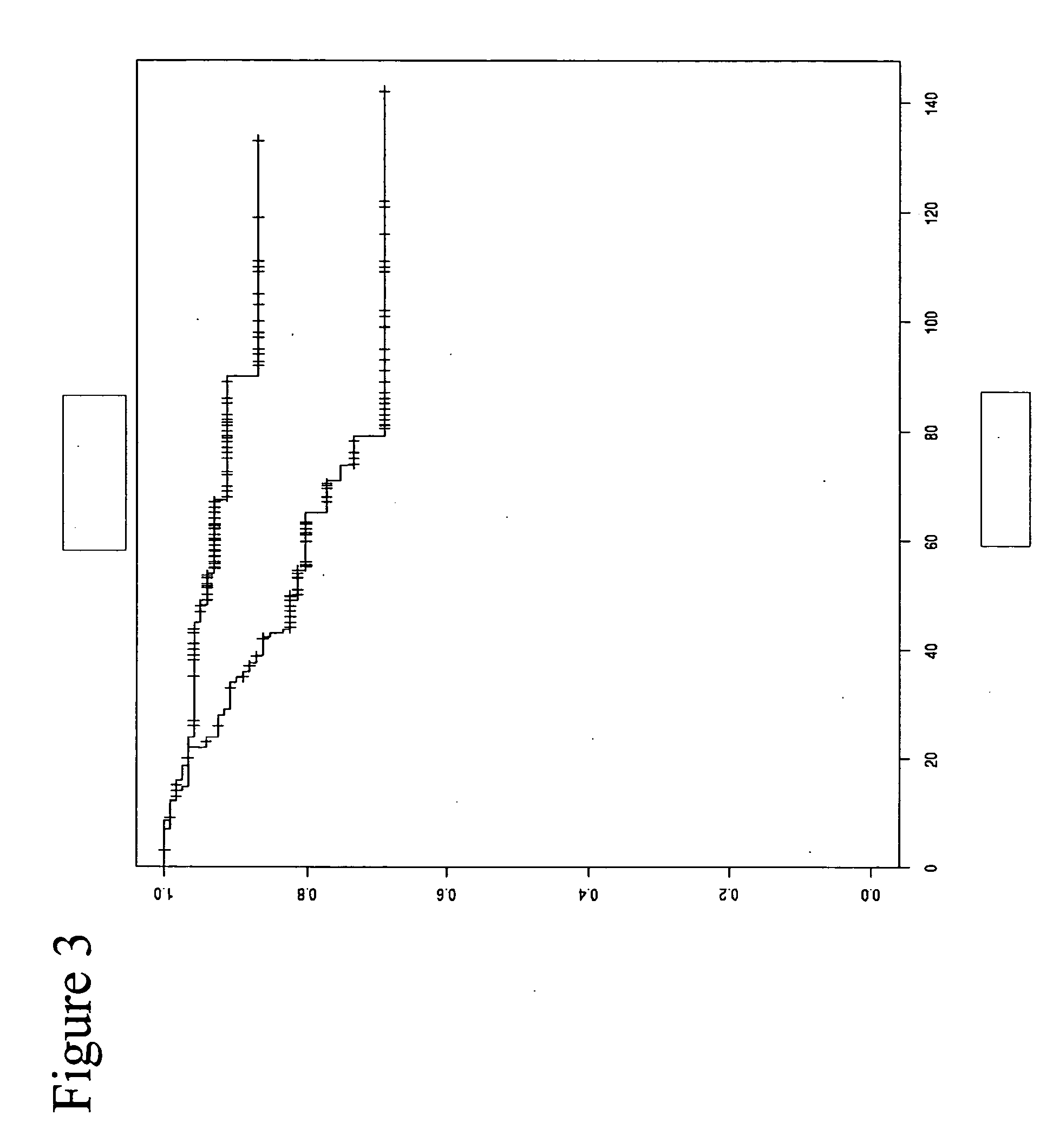
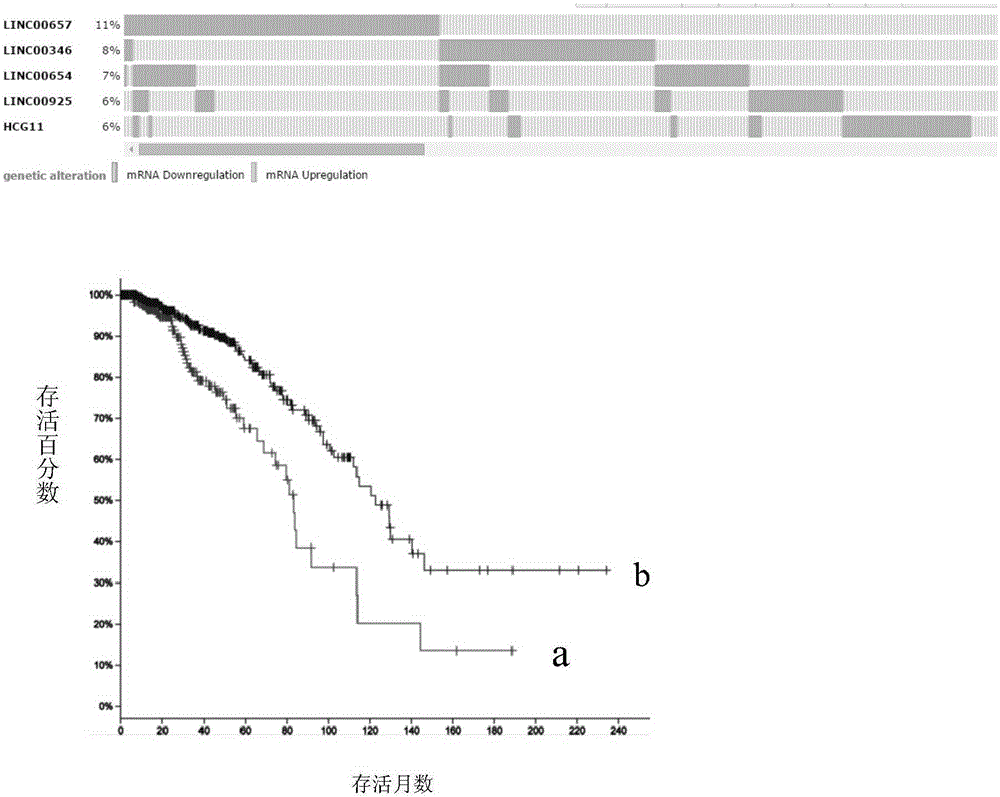
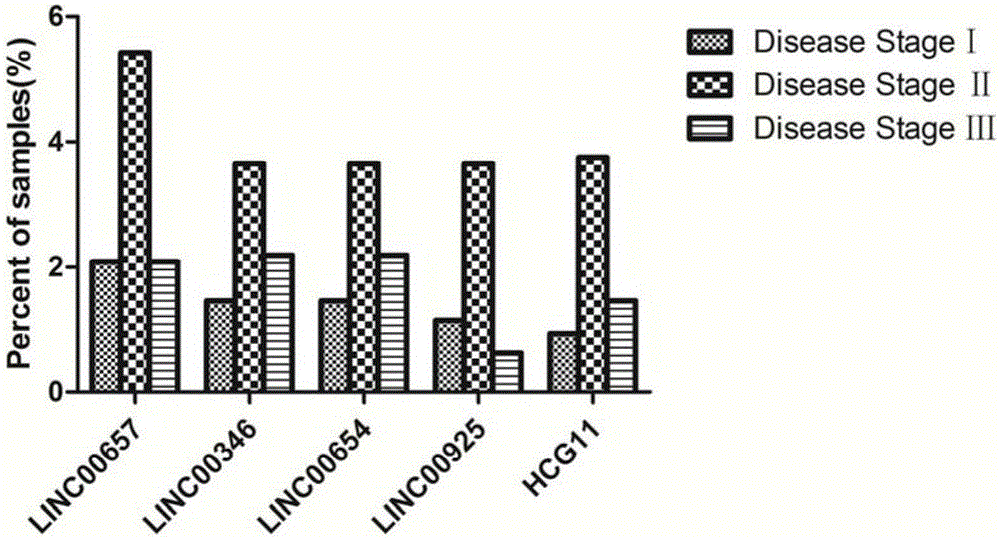
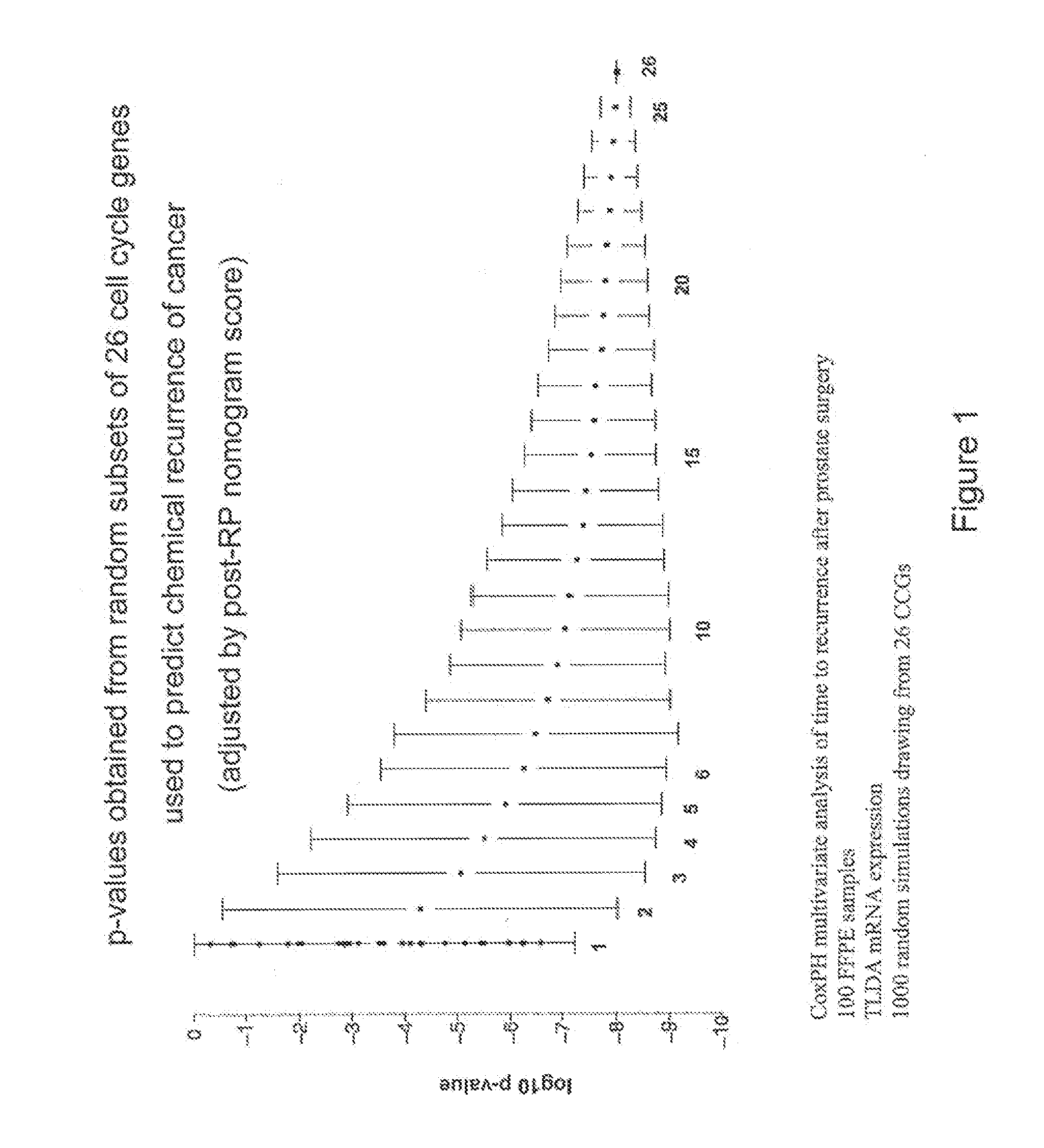
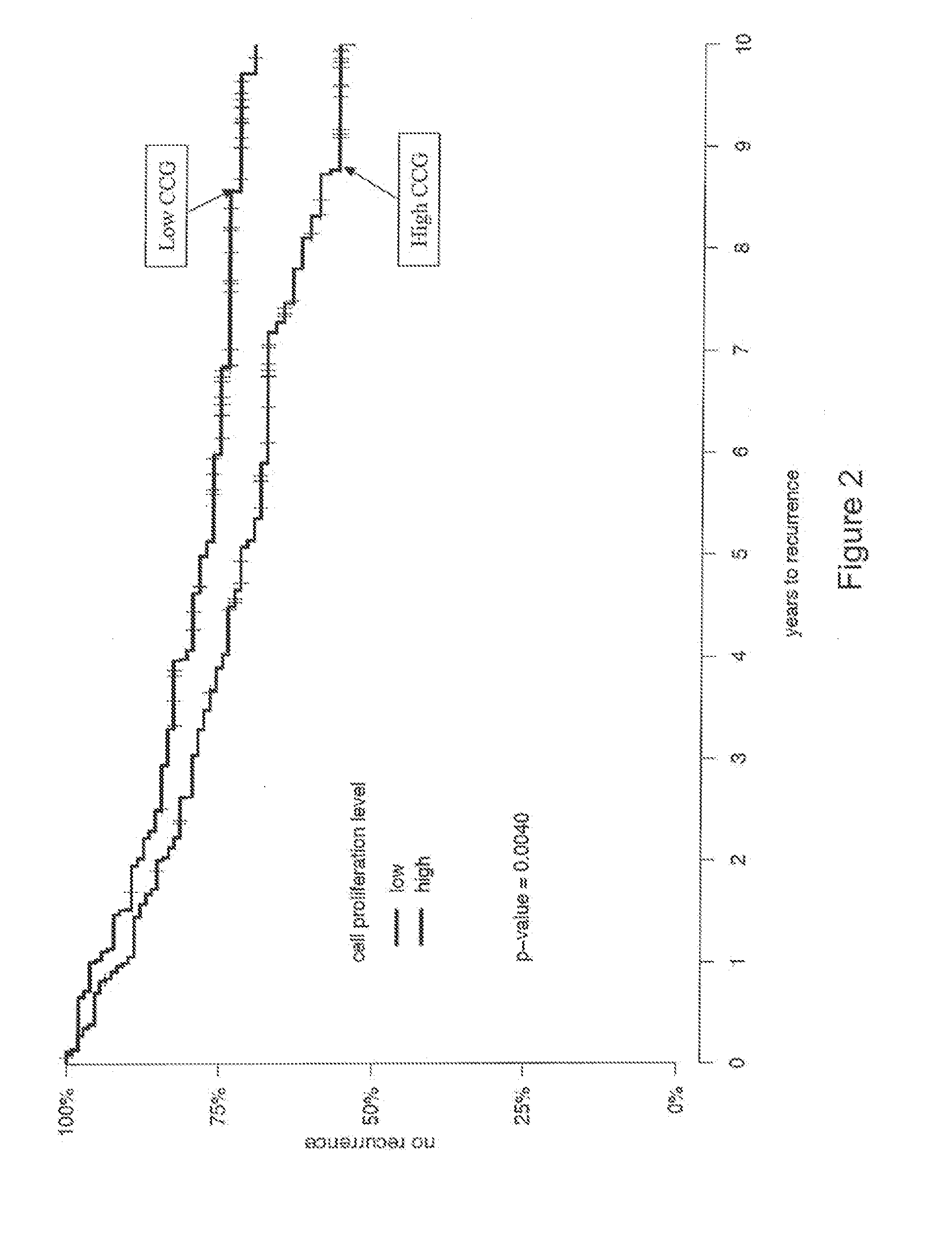
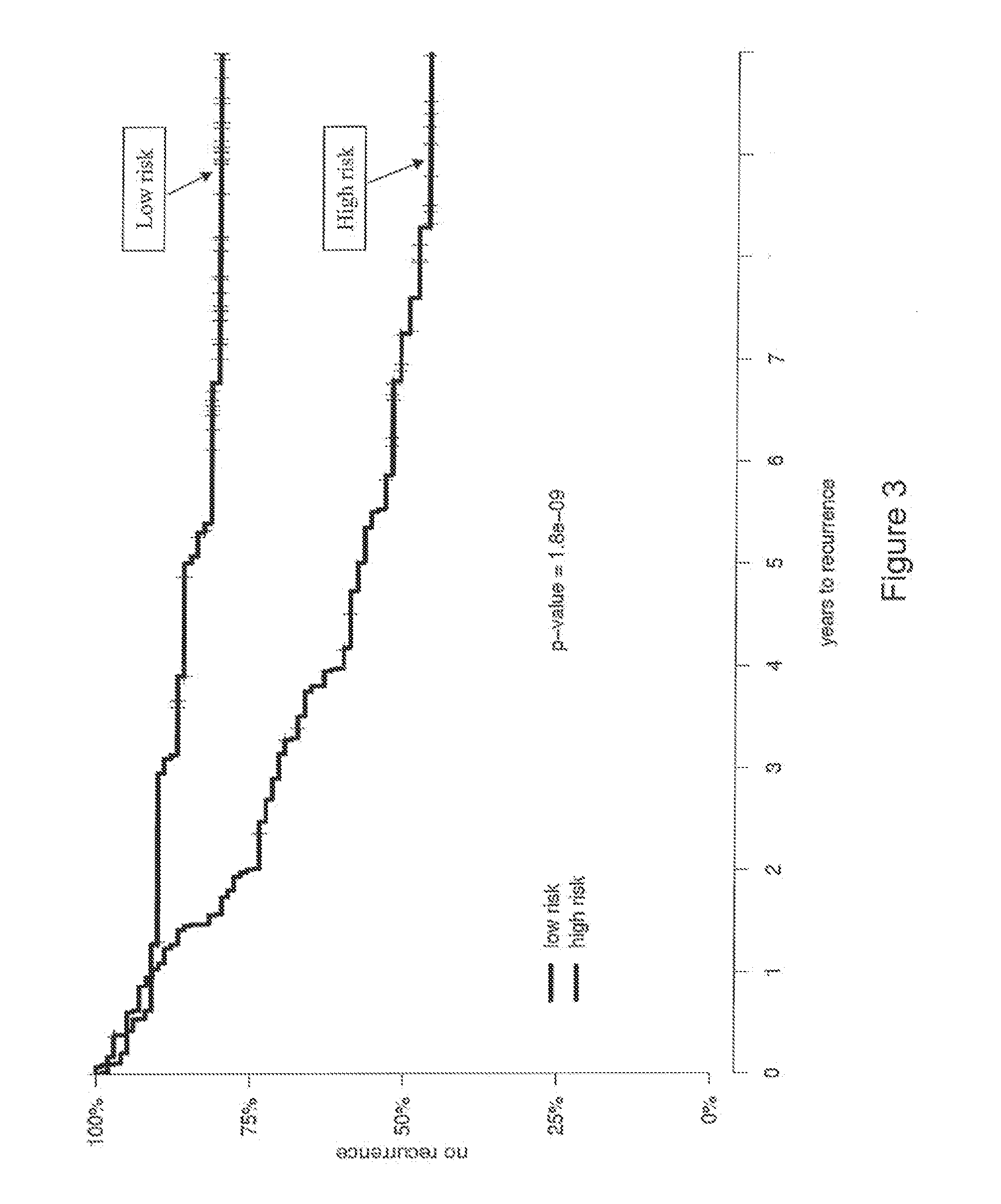
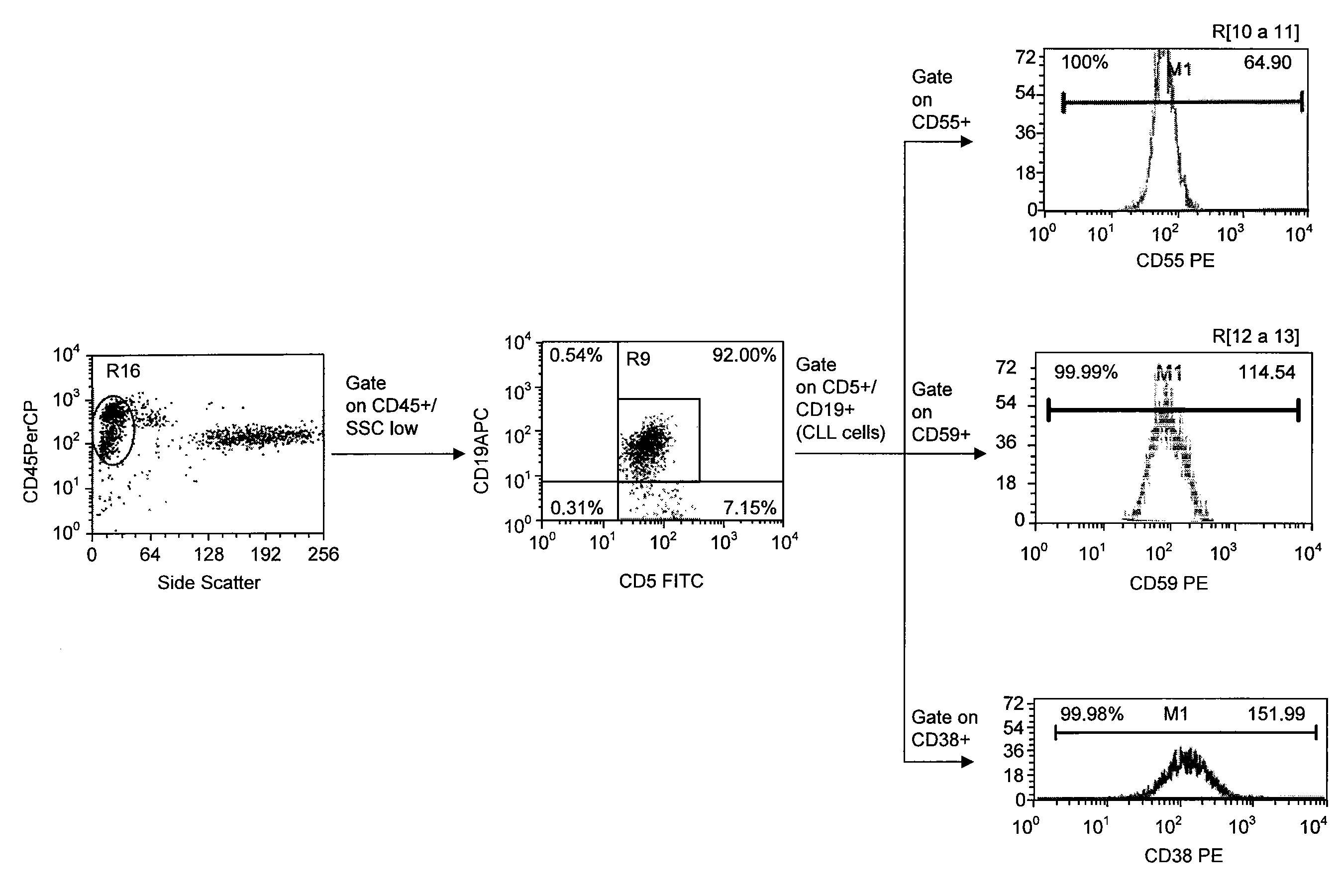
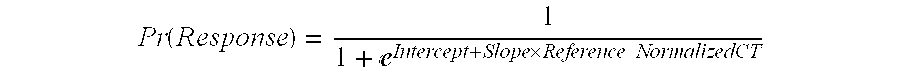Patents
Literature
156 results about "Prognosis biomarker" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
A prognostic biomarker helps indicate how a disease may develop in an individual when a disorder is already diagnosed. The presence or absence of a prognostic marker can be useful for the selection of patients for treatment but does not directly predict the response to a treatment.
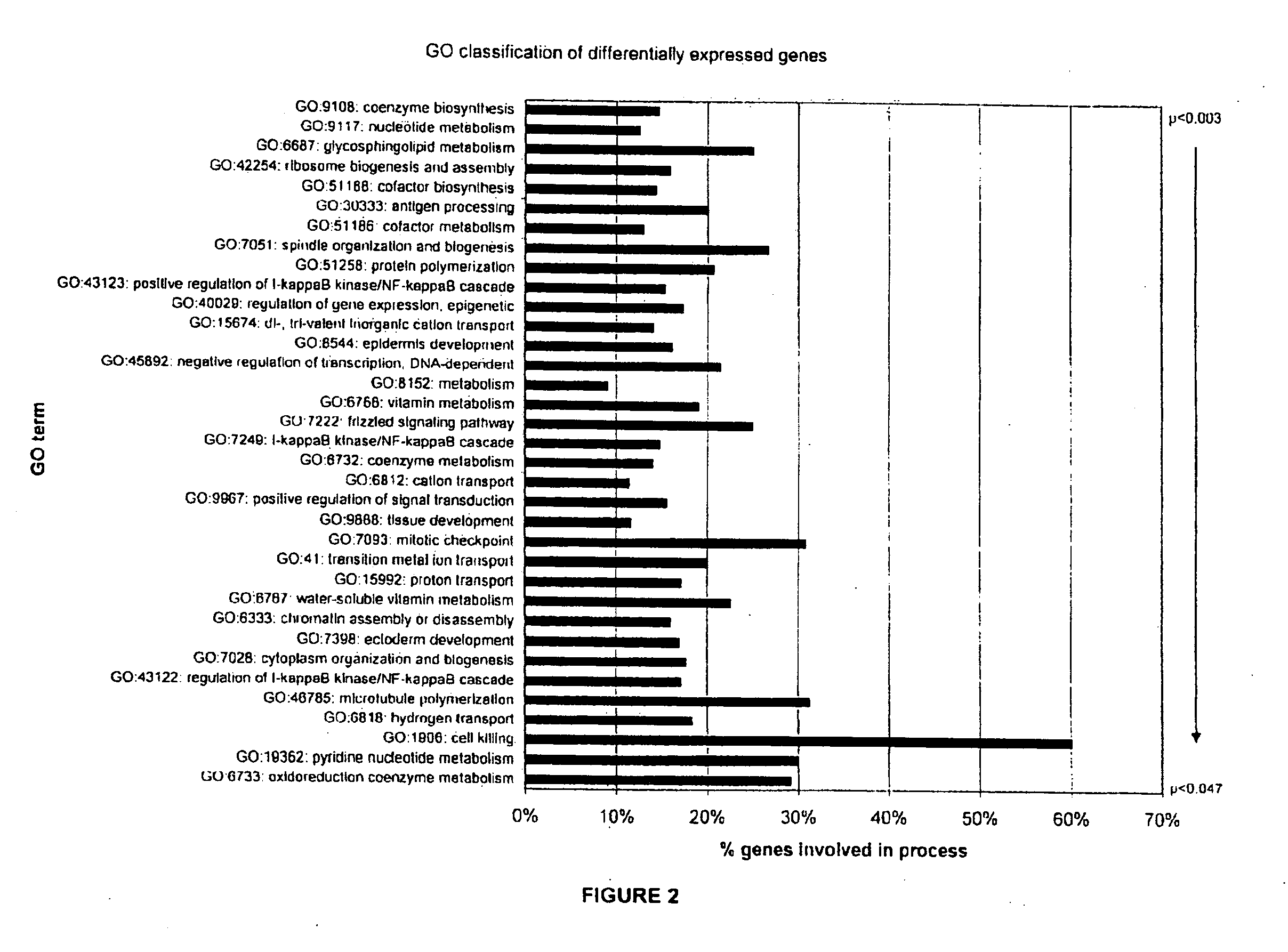
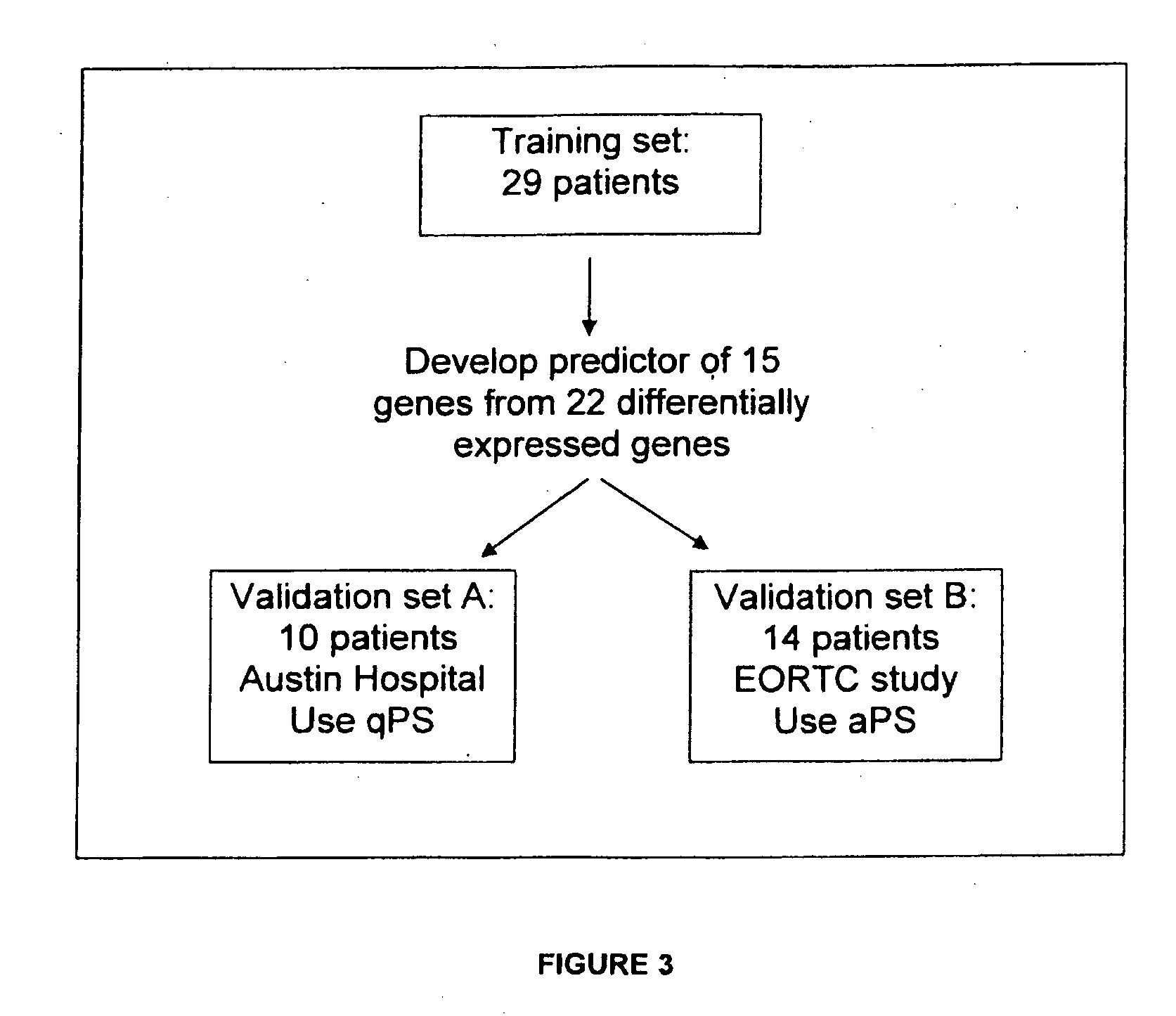
Gene expression profiling of EGFR positive cancer
InactiveUS20050019785A1Bioreactor/fermenter combinationsBiological substance pretreatmentsTissue sampleBiology
The present invention concerns prognostic markers associated with EGFR positive cancer. In particular, the invention concerns prognostic methods based on the molecular characterization of gene expression in paraffin-embedded, fixed tissue samples of EGFR-expressing cancer, which allow a physician to predict whether a patient is likely to respond well to treatment with an EGFR inhibitor.
Owner:GENOMIC HEALTH INC
Markers for colorectal cancer
InactiveUS20060188883A1Reduce and eliminate biological activityReduce expressionMicrobiological testing/measurementDisease diagnosisCancer cellBifunctional
Provided are previously uncharacterised markers of cancers, for example colorectal cancers, and uses of these as diagnostic and prognostic markers of cancers, and in particular colorectal cancers. The markers are SEQ ID NO: 1—hnRNP-K; SEQ ID NO:2—HMG-1; SEQ ID NO:3—proteasome subunit alpha type 1; SEQ ID NO:4—bifunctional purine biosynthesis protein; SEQ ID NO:5—ST11; SEQ ID NO:6—annex in IV; SEQ ID NO:7—60 kDa heat shock protein; SEQ ID NO:8—T complex protein 1 beta subunit; SEQ ID NO:9—T complex protein 1 epsilon subunit; SEQ ID NO: 10—mortalin; and SEQ ID NO: 11—TER-ATPase. The invention further provides related methods and materials for the use of the markers in therapeutic intervention in colorectal and other cancers e.g. to specifically target neoplastic cells without causing significant toxicity in healthy tissues, and to provide methods for the evaluation of the ability of candidate therapeutic compounds to modulate the biological activity of cancerous cells from the colon, rectum and other tissues.
Owner:AUVATION +2
Methods and compositions for diagnosis and prognosis of renal injury and renal failure
InactiveUS20120190051A1Eliminate needPeptide/protein ingredientsTransferrinsErythropoietin receptorFactor VIII vWF
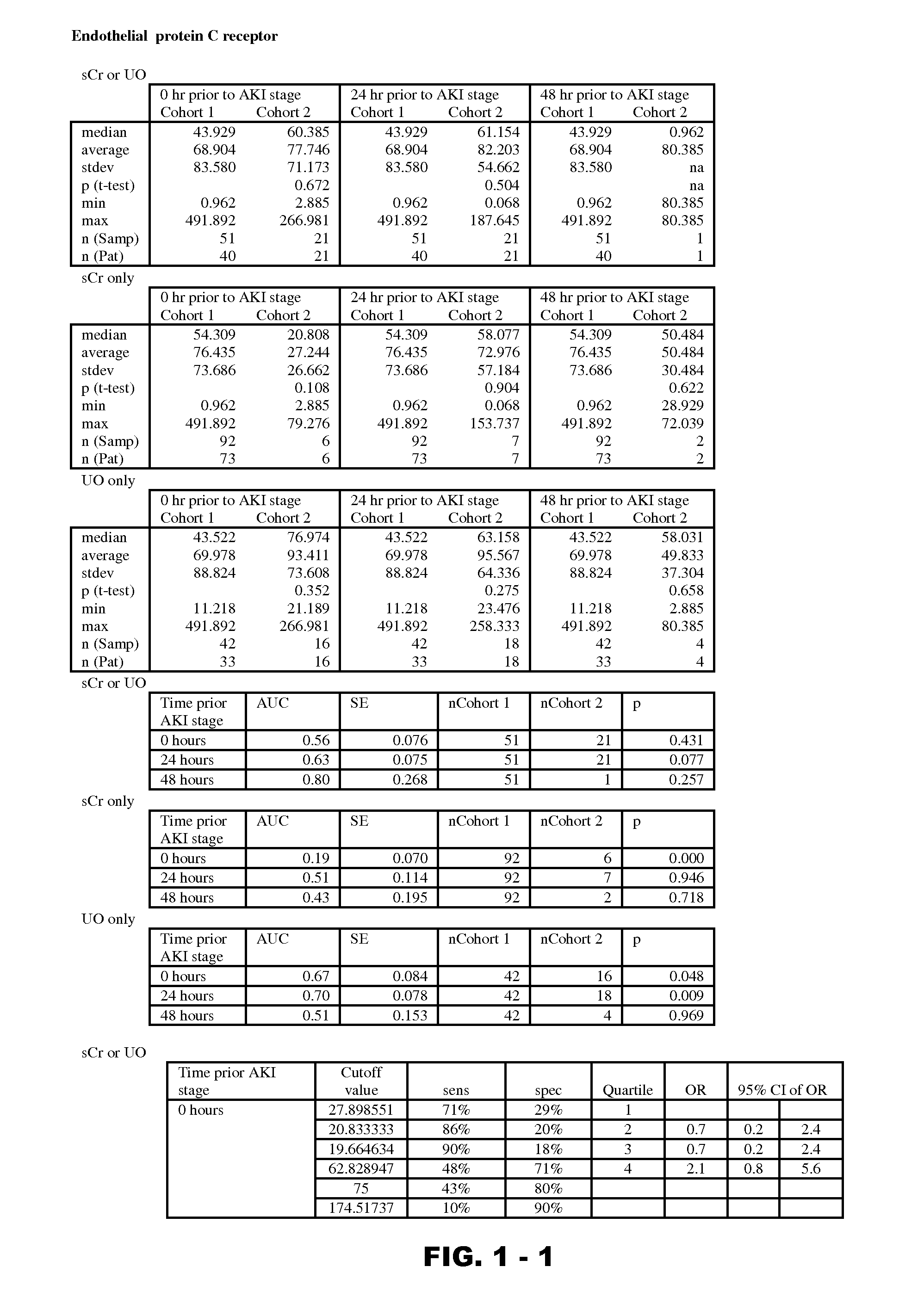
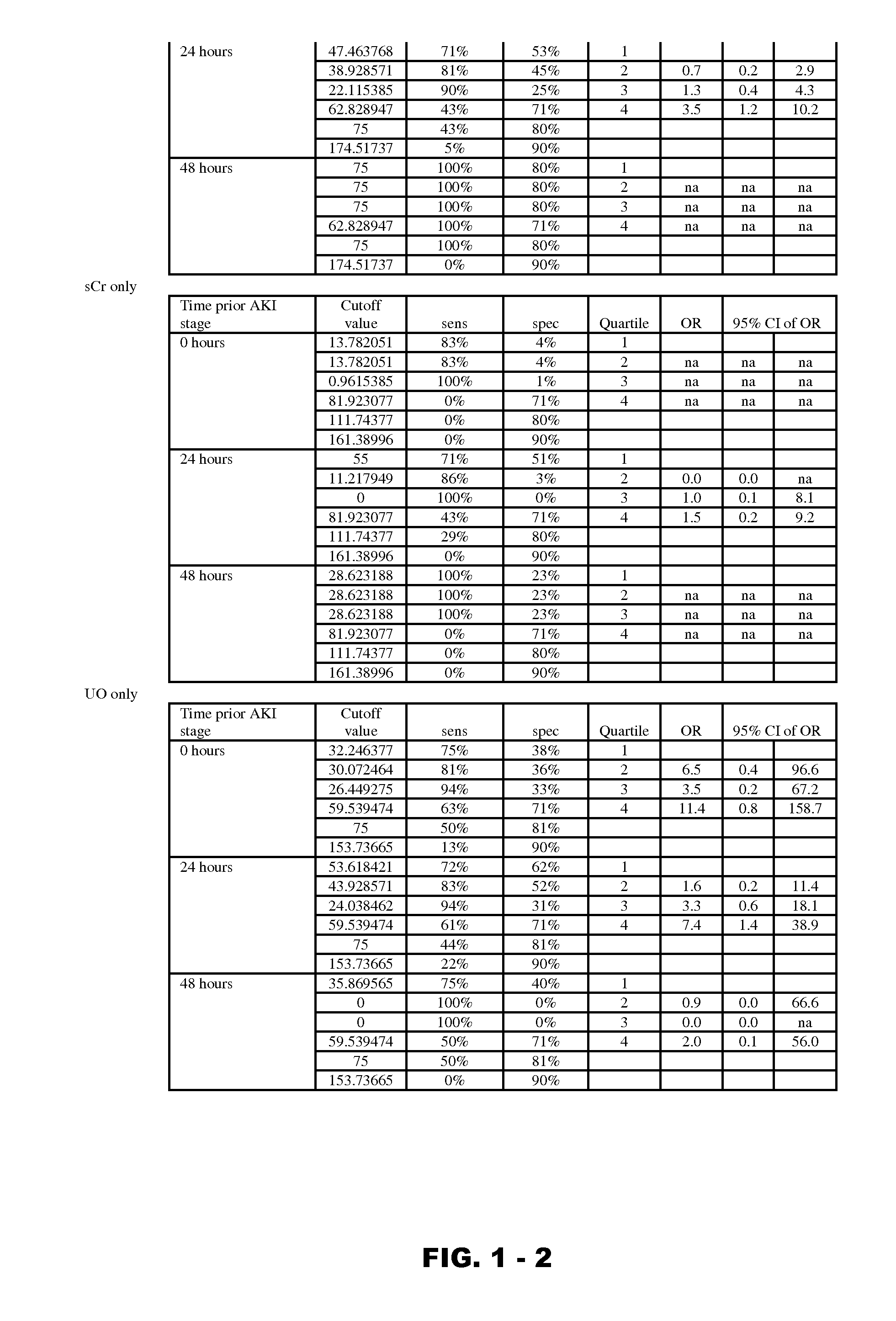
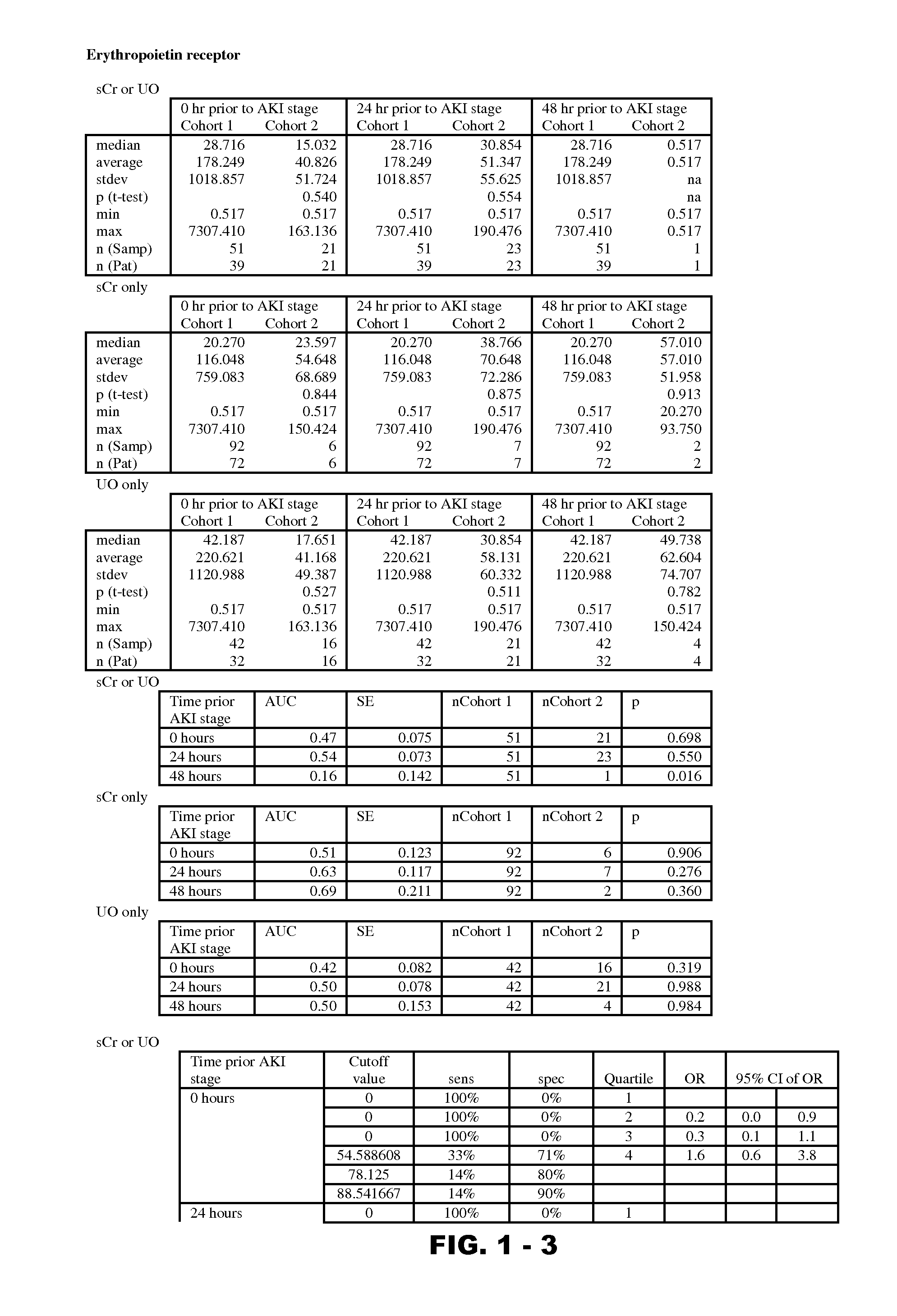
Disclosed are methods and compositions for monitoring, diagnosis, prognosis, and determination of treatment regimens in subjects suffering from or suspected of having a renal injury. In particular, disclosed are assays that detect one or more markers selected from the group consisting of Prostatic acid phosphatase, Lactotransfenin, Soluble erythropoietin receptor, Von Willebrand factor, Soluble endothelial protein C receptor, and Beta-2-glycoprotein 1 as diagnostic and prognostic biomarkers in renal injuries.
Owner:ASTUTE MEDICAL
Screening method of colorectal cancer treatment prognosis biomarkers
InactiveCN103324846AShorten screening timeReduce screening costsSpecial data processing applicationsDNA/RNA fragmentationKRASPrognosis biomarker
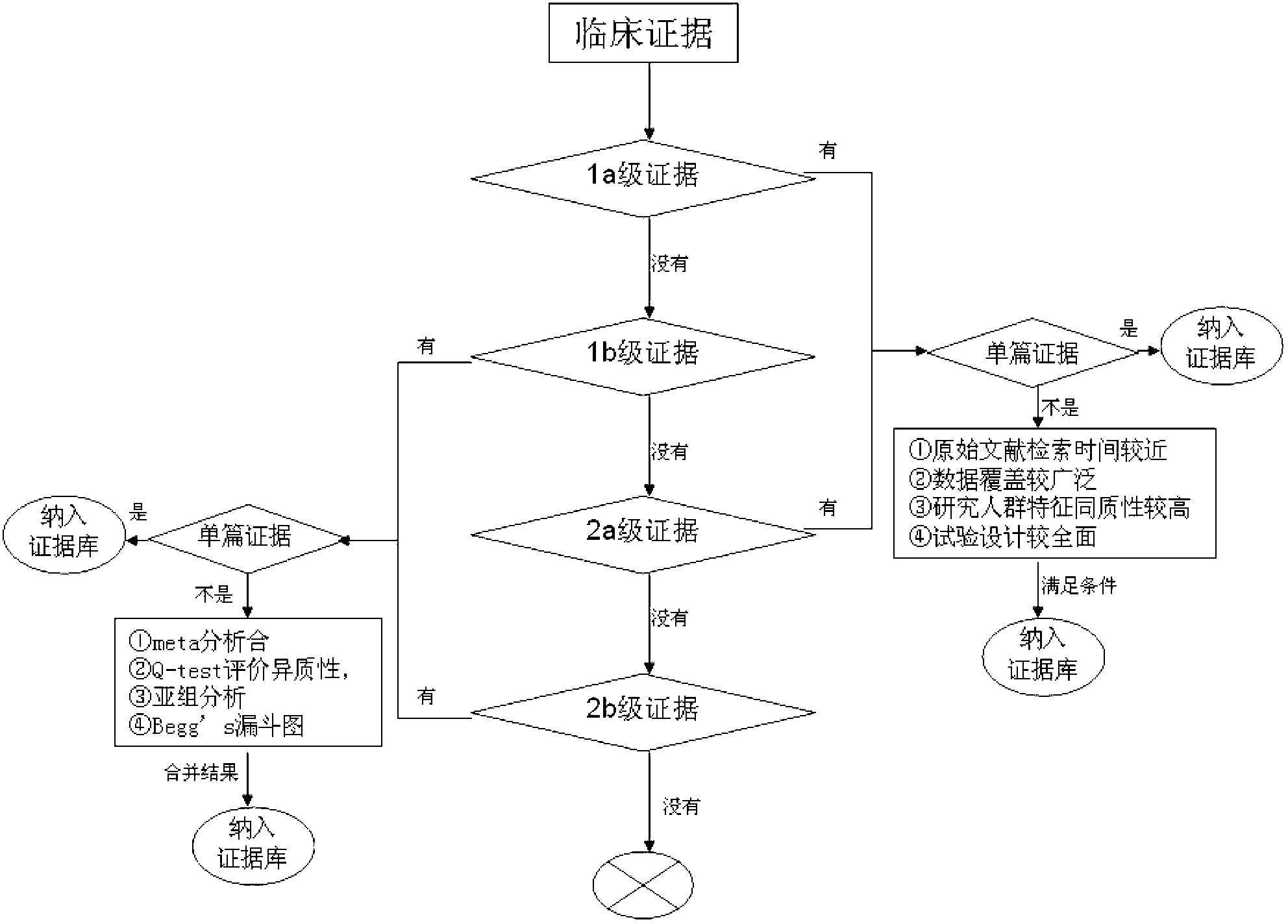
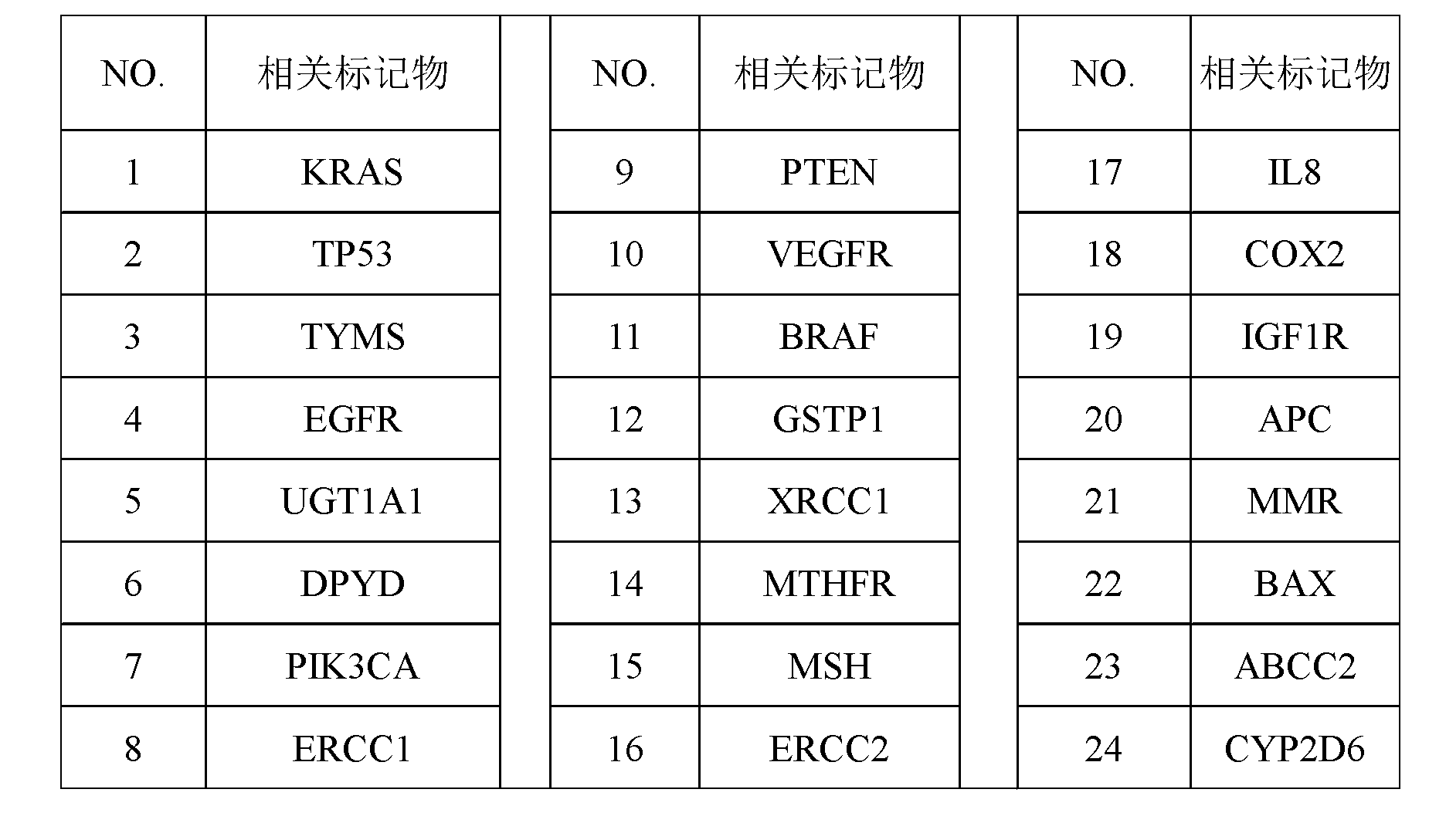
The invention discloses a screening method of colorectal cancer treatment prognosis biomarkers. The screening method includes the following steps of: (1) making a primary search strategy for screening the biomarkers; (2) making a literature adopting and exclusion criterion; (3) analyzing data and primarily screening the biomarkers; (4) making evident grades; (5) making a high quality evident evaluation criterion; (6) performing grading retrieval and quality evaluation on biomarker clinic evident; (7) performing data analyzing and statistics on the biomarker clinic evident; (8) screening the prognosis biomarkers. The invention further provides the colorectal cancer treatment prognosis biomarkers obtained through screening according to the screening method, and the prognosis biomarkers include KRAS, TYMS, TYMS, EGFR, UGT1A1*28, DPYD, PIK3CA, ERCC1, PTEN, VEGFR, BRAF, GST P1, XRCC1, MTHFR, MSI and the like.
Owner:浙江加州国际纳米技术研究院绍兴分院
Gene Expression Markers for Response to EGFR Inhibitor Drugs
InactiveUS20080176229A1Raise the possibilityMicrobiological testing/measurementFermentationPrognosis biomarkerPharmaceutical drug
The present invention concerns prognostic markers associated with cancer. In particular, the invention concerns prognostic methods based on the molecular characterization of gene expression in paraffin-embedded, fixed samples of cancer tissue, which allow a physician to predict whether a patient is likely to respond well to treatment with an EGFR inhibitor.
Owner:GENOMIC HEALTH INC +1
Gene expression profiling of EGFR positive cancer
InactiveUS8008003B2Bioreactor/fermenter combinationsBiological substance pretreatmentsPrognosis biomarkerTissue sample
The present invention concerns prognostic markers associated with EGFR positive cancer. In particular, the invention concerns prognostic methods based on the molecular characterization of gene expression in paraffin-embedded, fixed tissue samples of EGFR-expressing cancer, which allow a physician to predict whether a patient is likely to respond well to treatment with an EGFR inhibitor.
Owner:GENOMIC HEALTH INC
Interventricular delay as a prognostic marker for reverse remodeling outcome from cardiac resynchronization therapy
InactiveUS20070239037A1Increase geometryFunction increaseInternal electrodesCatheterLeft ventricular sizeMortality rate
In heart failure (HF) patients diagnosed according to the New York Heart Association (NYHA) rating scale as either Class III or Class IV, with QRS duration ≧120 ms, left ventricular (LV) EF≦35% (when in sinus rhythm), cardiac resynchronization therapy (CRT) can predict sustained improvement in at least LV structure and function. The knowledge of aetiology of a subject's HF status and of a few simple echocardiographic characteristics provides useful information as to whether such patients and undergo significant and beneficial reverse remodelling of the LV in particular and, in the long term can be expected to experience an improvement in all-cause mortality, for example such patients can be reasonably expected to survive and enjoy a relatively enhanced quality of life (QOL). A patient who qualifies according to the invention can be termed a reverse remodelling responder (RRR) or the like.
Owner:MEDTRONIC INC
Genetic amplification of IQGAP1 in cancer
ActiveUS9157123B2Diminish invasivenessReduce spreadOrganic active ingredientsSugar derivativesCell invasionFollicular thyroid cancer
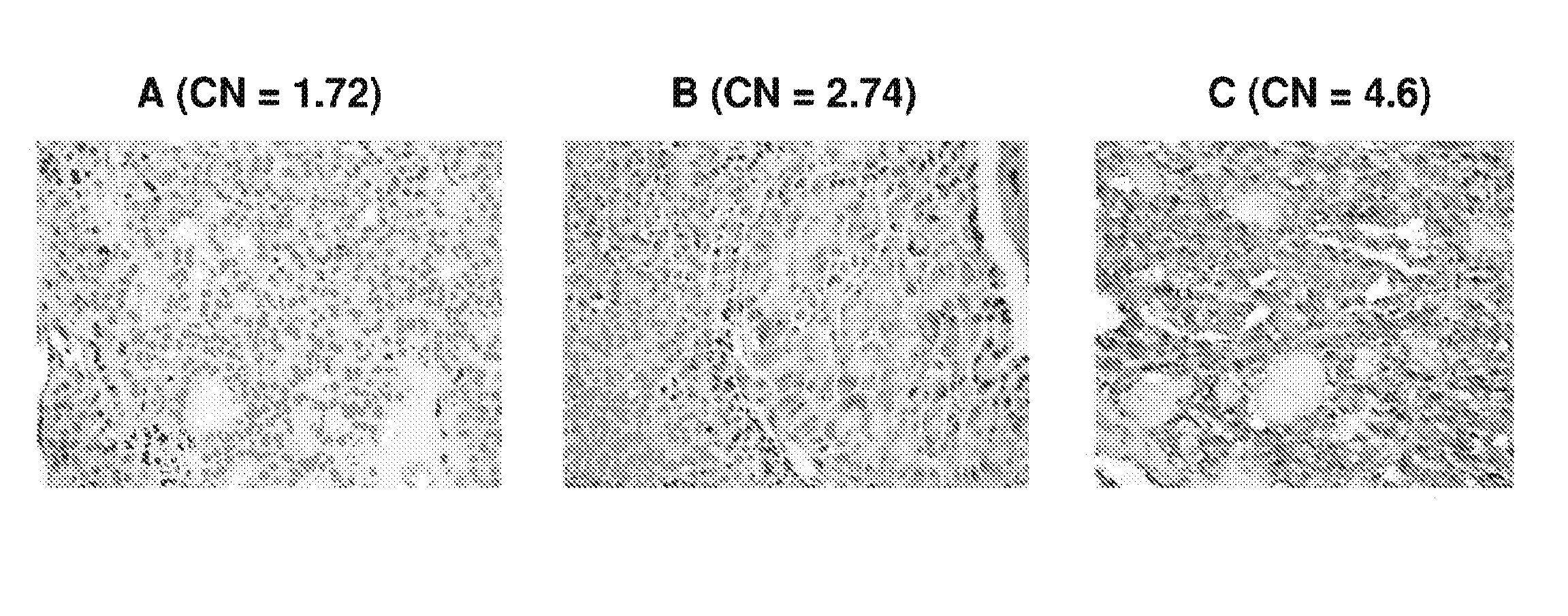
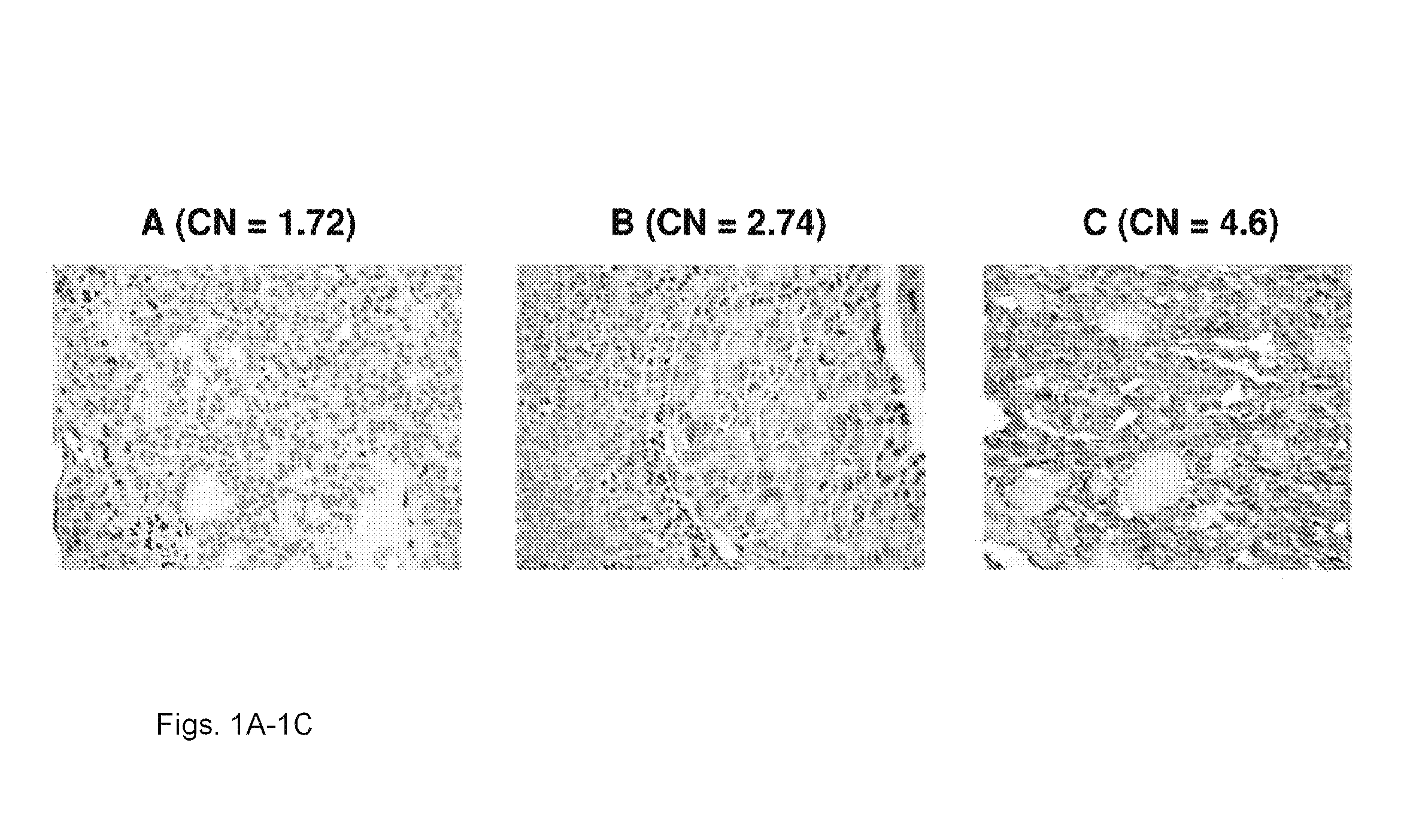
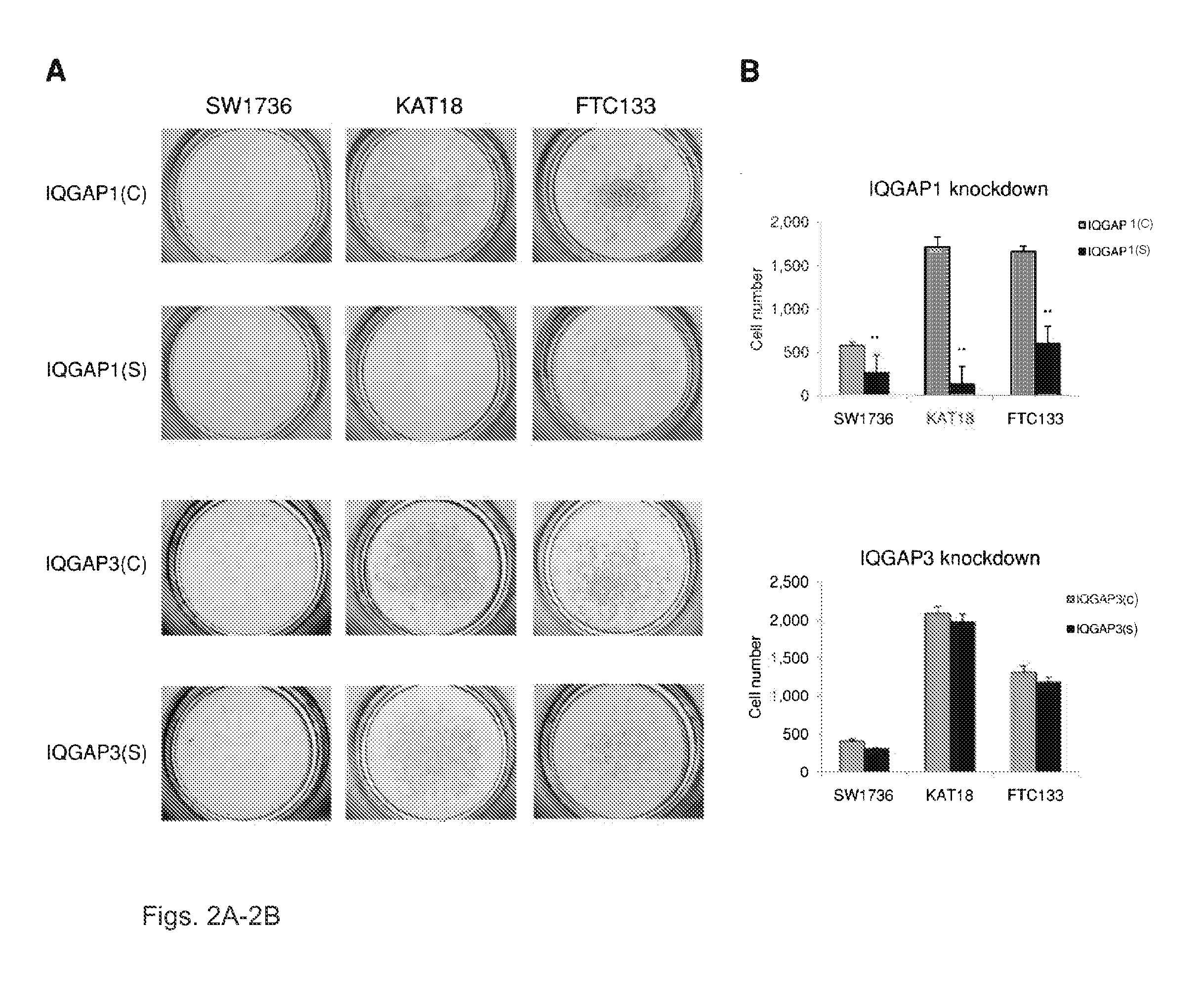
We examined IQGAP1 copy gain and its relationship with clinicopathologic outcomes of thyroid cancer and investigated its role in cell invasion and molecules involved in the process. We found IQGAP1 copy number (CN) gain ?3 in 1 of 30 (3%) of benign thyroid tumor, 24 of 74 (32%) follicular variant papillary thyroid cancer (FVPTC), 44 of 107 (41%) follicular thyroid cancer (FTC), 8 of 16 (50%) tall cell papillary thyroid cancer (PTC), and 27 of 41 (66%) anaplastic thyroid cancer, in increasing order of invasiveness of these tumors. A similar tumor distribution trend of CN ?4 was also seen. IQGAP1 copy gain was positively correlated with IQGAP1 protein expression. It was significantly associated with extrathyroidal and vascular invasion of FVPTC and FTC and, remarkably, a 50%-60% rate of multifocality and recurrence of BRAF mutation-positive PTC (P=0.01 and 0.02, respectively). The siRNA knock-down of IQGAP1 dramatically inhibited thyroid cancer cell invasion and colony formation. Co-immunoprecipitation assay showed direct interaction of IQGAP1 with E-cadherin, a known invasion-suppressing molecule, which was upregulated when IQGAP1 was knocked down. IQGAP1, through genetic copy gain, plays an important role in the invasiveness of thyroid cancer and represents a useful prognostic marker and therapeutic target for this and other cancers.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
Prognosis prediction for melanoma cancer
ActiveUS20100136553A1Effective individual testImprove forecast accuracySugar derivativesHealth-index calculationProtein markersPrognostic signature
The invention relates to prognostic markers and prognostic signatures, and compositions and methods for determining the prognosis of cancer in a patient, particularly for melanoma. Specifically, the invention relates to the use of genetic and protein markers for the prediction of the risk of progression of a cancer, such as melanoma, based on markers and signatures of markers. In various aspects, the invention provides methods, compositions, kits, and devices based on prognostic cancer markers, specifically melanoma prognostic markers, to aid in the prognosis and treatment of cancer.
Owner:PACIFIC EDGE +1
Methods and compositions for diagnosis and prognosis of renal injury and renal failure
InactiveUS9229010B2Peptide/protein ingredientsMammal material medical ingredientsFactor VIII vWFErythropoietin receptor
Disclosed are methods and compositions for monitoring, diagnosis, prognosis, and determination of treatment regimens in subjects suffering from or suspected of having a renal injury. In particular, disclosed are assays that detect one or more markers selected from the group consisting of Prostatic acid phosphatase, Lactotransfenin, Soluble erythropoietin receptor, Von Willebrand factor, Soluble endothelial protein C receptor, and Beta-2-glycoprotein 1 as diagnostic and prognostic biomarkers in renal injuries.
Owner:ASTUTE MEDICAL
Method for establishing pancreatic cancer miRNA prognosis model and screening targeted gene
PendingCN112391470AMicrobiological testing/measurementBiostatisticsPancreas CancersTreatment targets
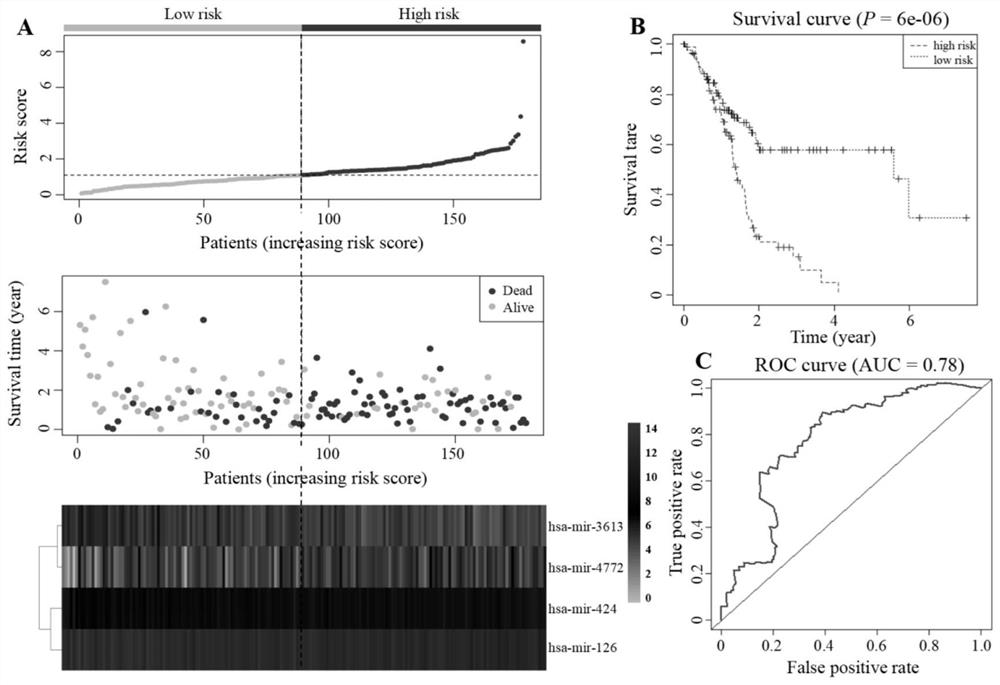
The invention discloses a method for establishing a pancreatic cancer miRNA prognosis model and screening a targeted gene. The model comprises hsa-mir-424, hsa-mir-126, hsa-mir-3613 and hsa-mir-4772,nine key genes are identified, and the nine key genes comprise MMP14, ITGA2, THBS2, COL1A1, COL3A1, COL11A1, COL6A3, COL12A1 and COL5A2. According to the method for establishing the pancreatic cancermiRNA prognosis model, by means of TCGA and GEO databases, data is subjected to multi-step analysis through a plurality of R language installation packages and combines with clinical information, a Cox proportional risk regression model is established to search for prognosis biomarkers, the targeted gene of miRNA is predicted, the key genes related to pancreatic cancer are found out by utilizing Cytoscape, and related molecular functions and action mechanisms of the key genes are predicted through KEGG and GO analysis so as to search for new treatment targets and prognosis markers of pancreatic cancer patients.
Owner:GUANGDONG MEDICAL UNIV +1
Gene sets and scoring model for evaluating tumor microenvironment, and application of gene sets
ActiveCN112133365ASignificant prognostic differenceGood forecastMicrobiological testing/measurementBiostatisticsFavorable prognosisPrognosis biomarker
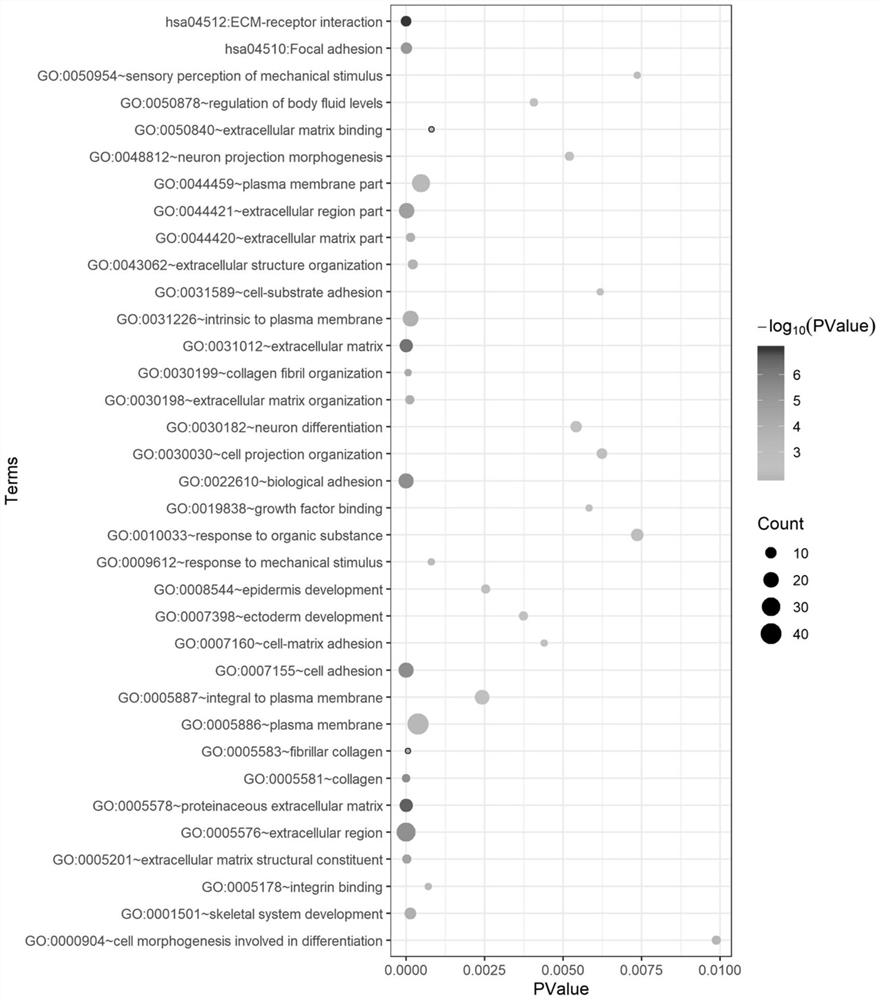
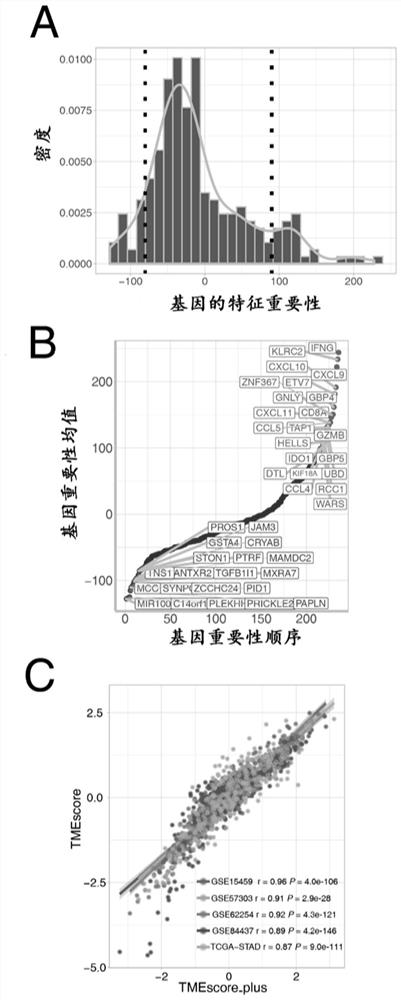
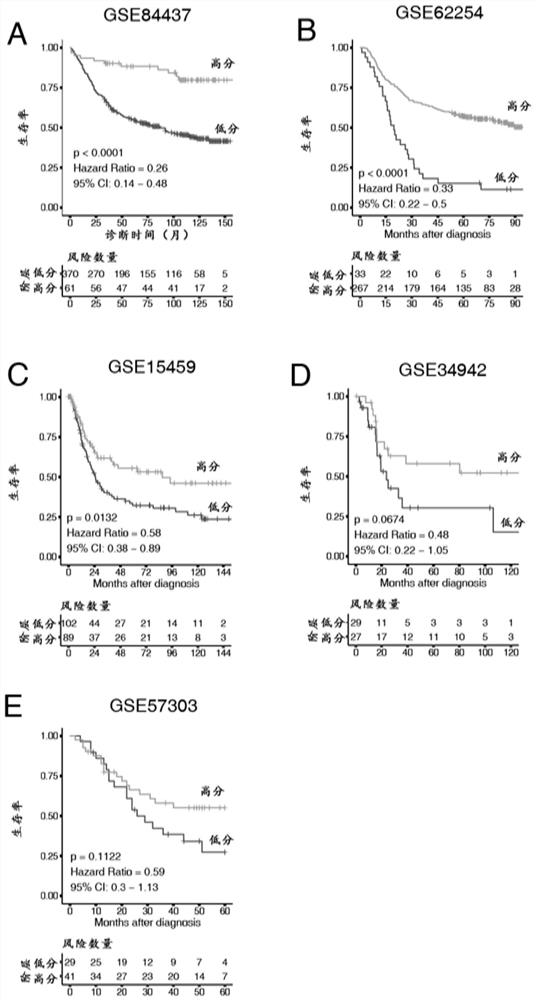
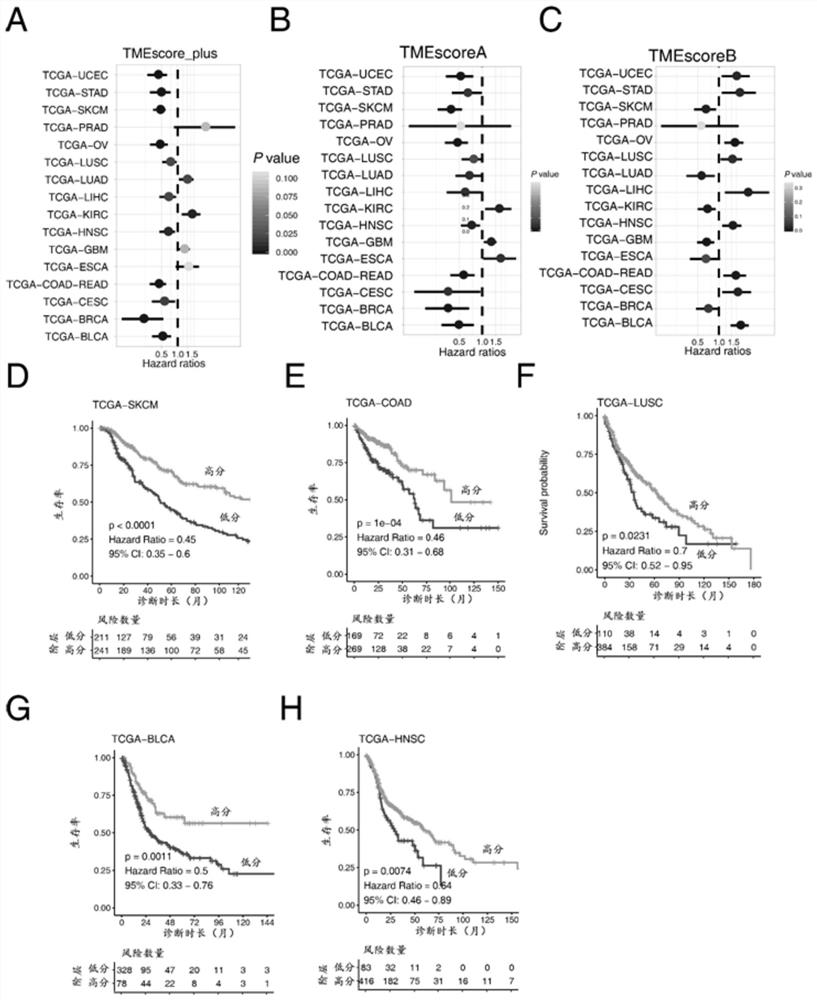
The invention discloses gene sets and a scoring model for evaluating a tumor microenvironment, and application of the gene sets, and belongs to the field of biological information. It is proved for the first time that the gastric cancer has remarkable tumor microenvironment typing, and prognosis difference of gastric cancer patients with different typing is remarkable. According to a constructionmethod of the multi-gene scoring model for evaluating the gastric cancer tumor microenvironment, 44 gene sets capable of representing different microenvironment types are screened out, and the prediction efficiency of the tumor microenvironment multi-gene scoring model corresponding to the gene sets is remarkably better than that of other existing models; and the model is not only a prognostic marker of a gastric cancer patient, but also has a quite remarkable prediction effect in pan-cancer. The model provided by the invention not only can be used as a good prognosis biomarker, but also can be used for predicting checkpoint inhibitor immunotherapy response of various tumors by effectively evaluating the tumor microenvironment, and has important significance and clinical application valuefor better screening immunotherapy benefiting people.
Owner:广东隆宇医疗科技有限公司
Prognostic Marker for Endometrial Carcinoma
InactiveUS20110217701A1Microbiological testing/measurementDisease diagnosisNon small lung cancerRegimen
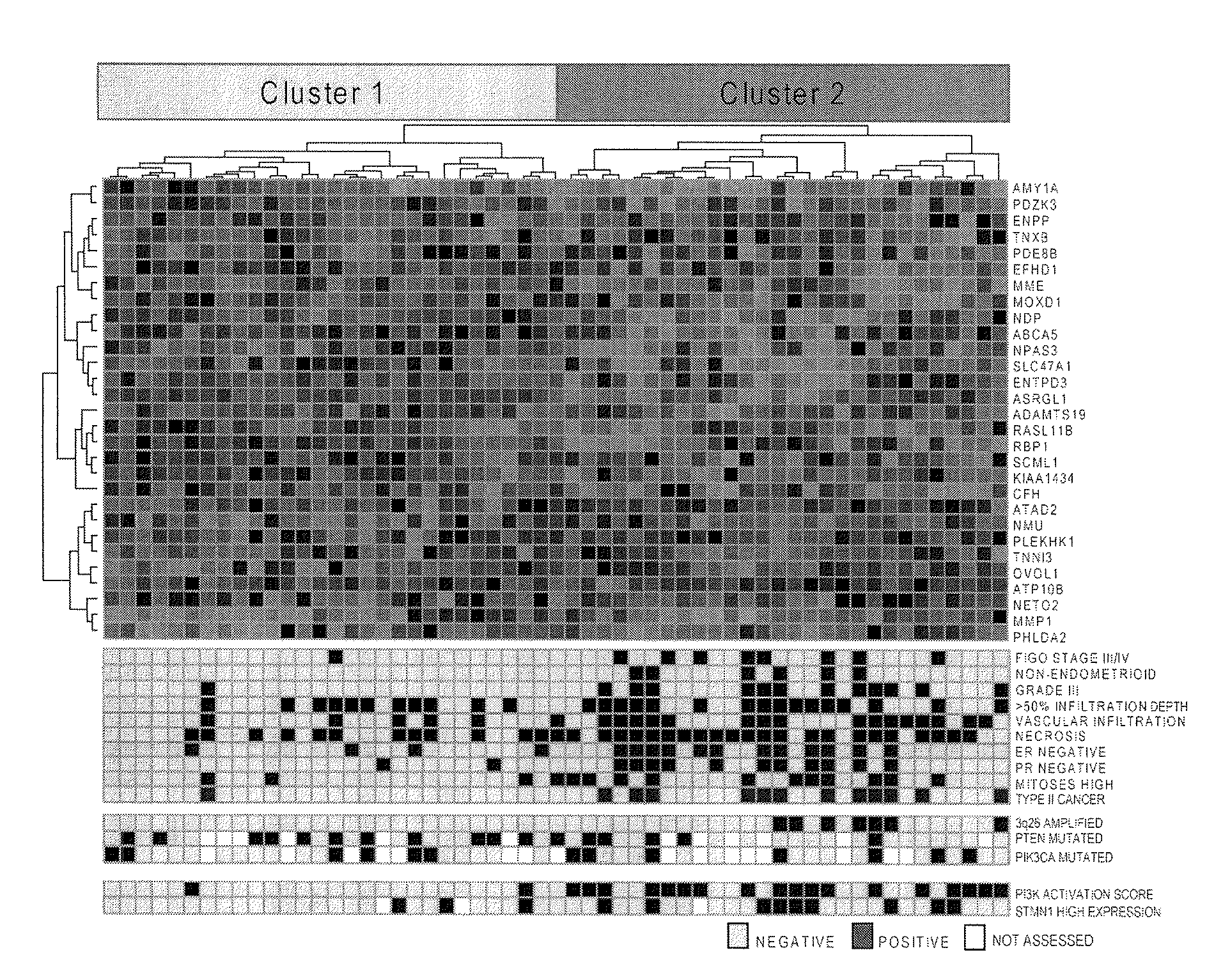
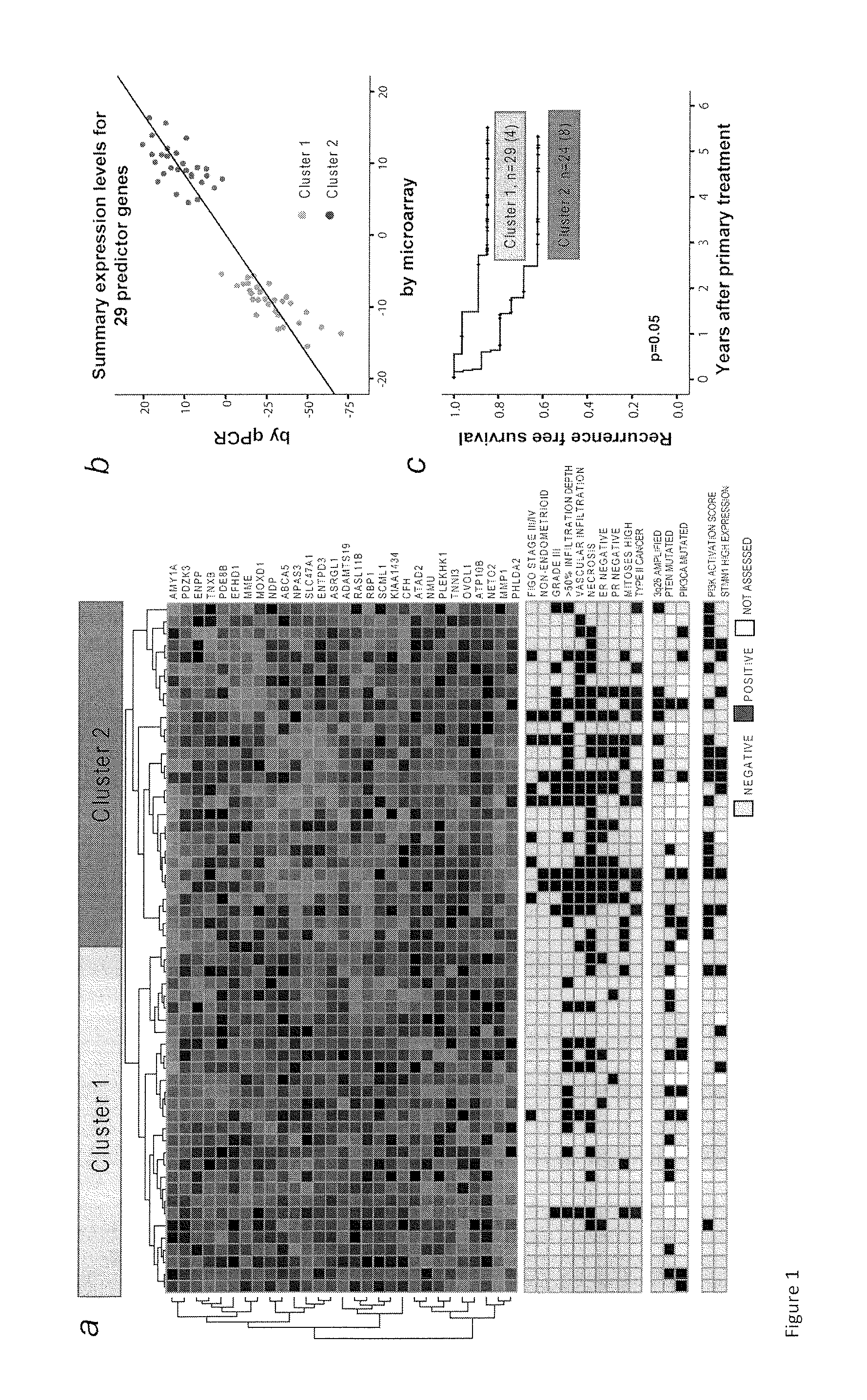
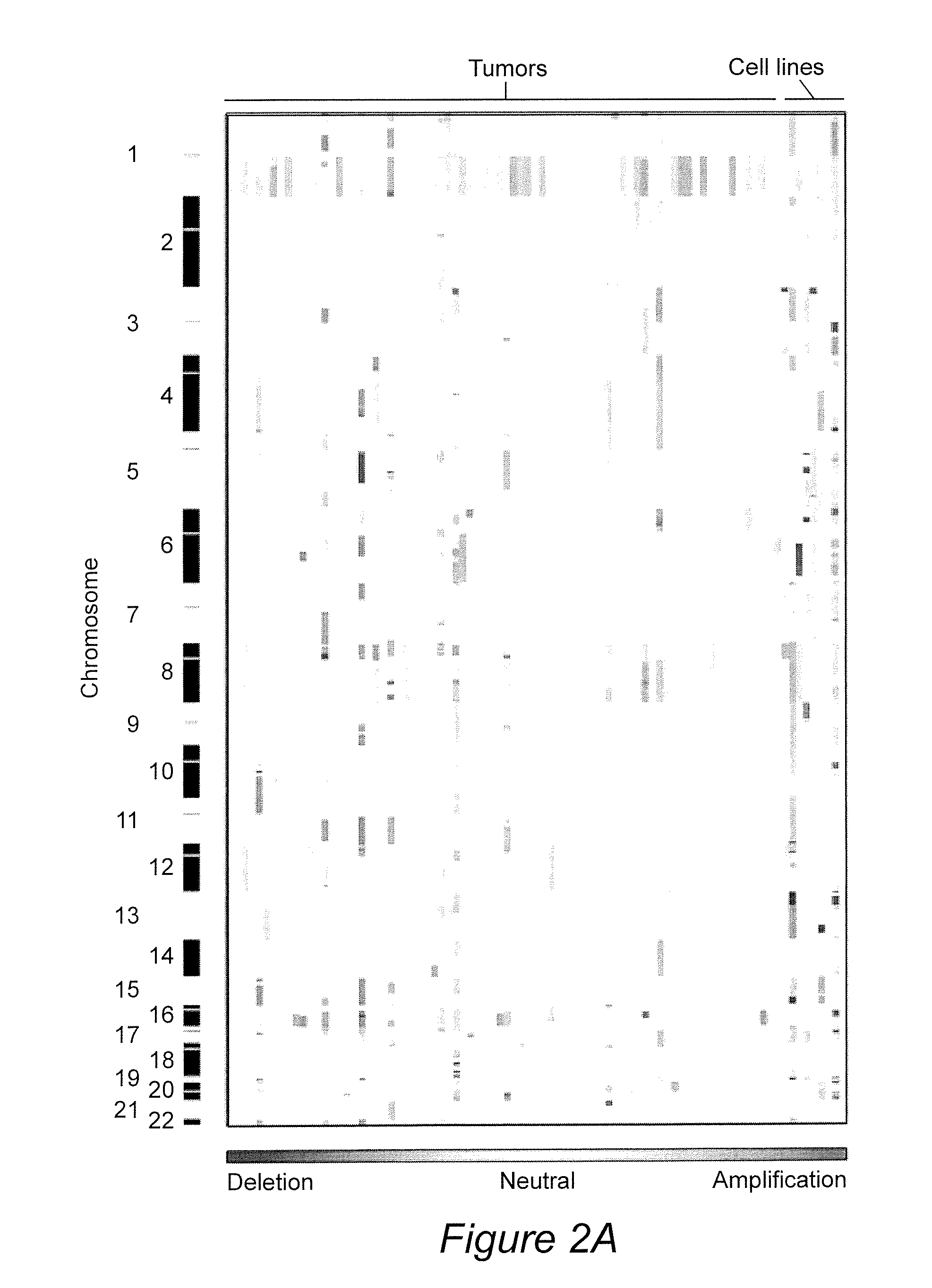
The present invention relates to a method for diagnosis of different stages of endometrial cancer in an individual. Further, the present invention relates to a method for evaluating the probability of survival for an individual suffering from endometrial carcinoma. In another aspect, the present invention relates to the stratification of therapy regimen of endometrial tumor, ovarian cancer, breast cancer, non-small lung cancer or hormone refractory prostate cancer therapy in an individual or monitoring therapeutic efficacy in an individual suffering from the same based on the expression status of STMN1 gene or protein. Moreover, the present invention relates to a kit for use in any of the above referenced methods comprising a means for determining amplifications and deletions of chromosomal regions 3q26.32 and 12p12.1, determining alterations of the gene expression profile of the genes (gene signature): upregulation of the genes PLEKHK1, ATP10B, NMU, MMP1, ATAD2, NETO2, TNNI3, PHLDA2, OVOL1 and down-regulation of the genes: NDP, KIAA1434, MME, CFH, MOXD1, SLC47A1, RBP1, PDE8B, ASRGL1, ADAMTS19, EFHD1, ABCA5, NPAS3, SCML1, TNXB, ENTPD3, AMY1A, ENPP, RASL11B, PDZK3, or the expression status of the STMN1 gene or protein, respectively. Finally, the present invention provides a method for predicting the response to taxanes in an individual suffering from a disease treated with the taxanes based on the expression status of the STMN1 gene or protein.
Owner:BERGEN TEKNOLOGIOVERFORING
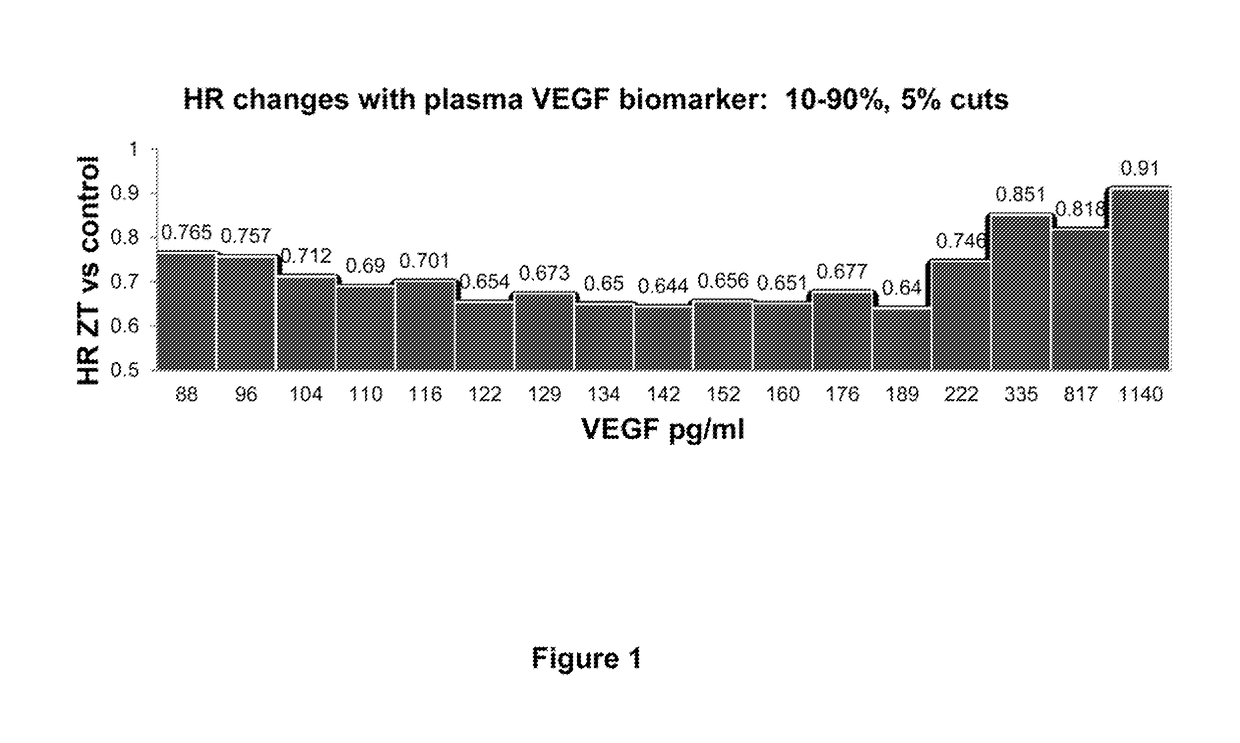
Predictive and prognostic biomarkers related to Anti-angiogenic therapy of metastatic colorectal cancer
ActiveUS20170281725A1Effective treatmentPeptide/protein ingredientsDisease diagnosisFAVORABLE RESPONSEPrognosis biomarker
The present invention provides methods for treating metastatic cancer comprising identifying subjects who will respond favorably to anti-VEGF therapy. According to certain aspects of the invention, subjects are identified based on their expression level of one or more predictive biomarkers. Favorable response to anti-VEGF therapy is indicated by high expression levels of certain biomarkers or by low expression levels of certain biomarkers. An exemplary predictive biomarker is VEGF-A. Also disclosed herein are prognostic biomarkers useful for identifying cancer-bearing subjects who are expected to have better relative survival outcomes.
Owner:REGENERON PHARM INC
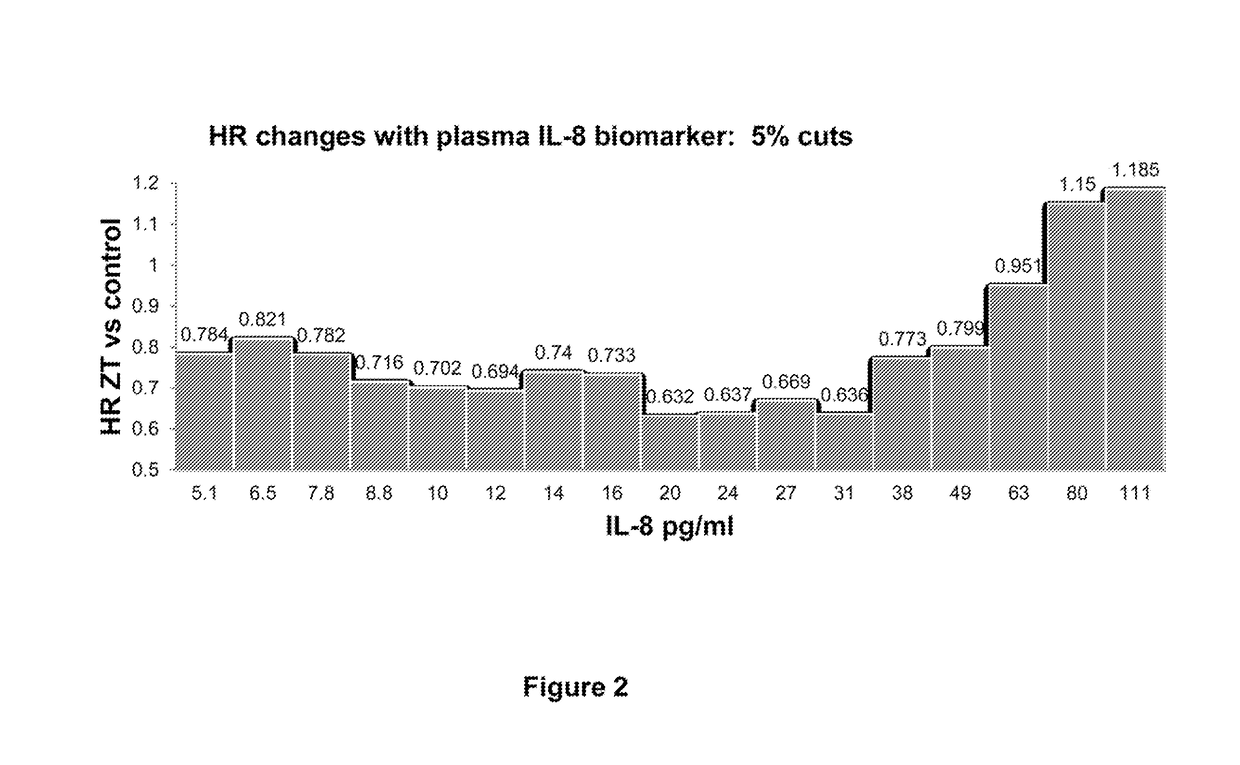
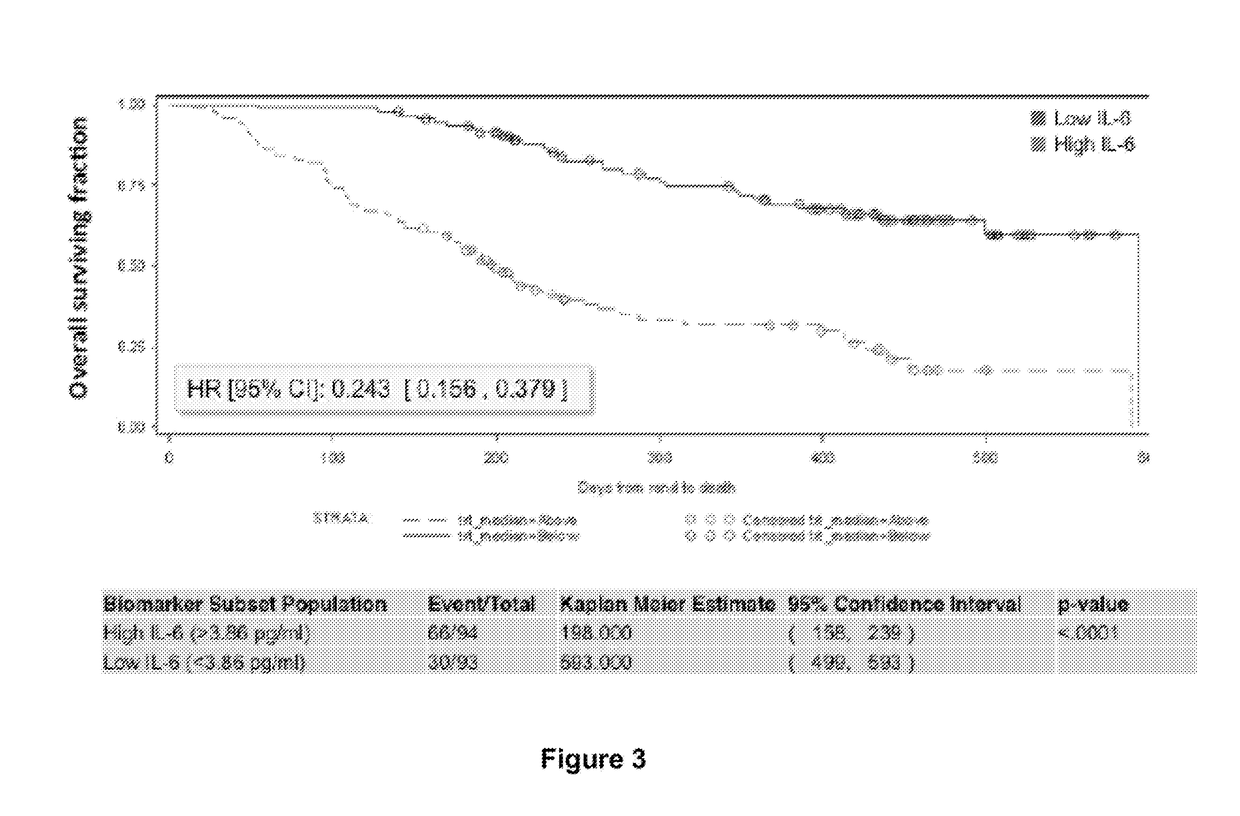
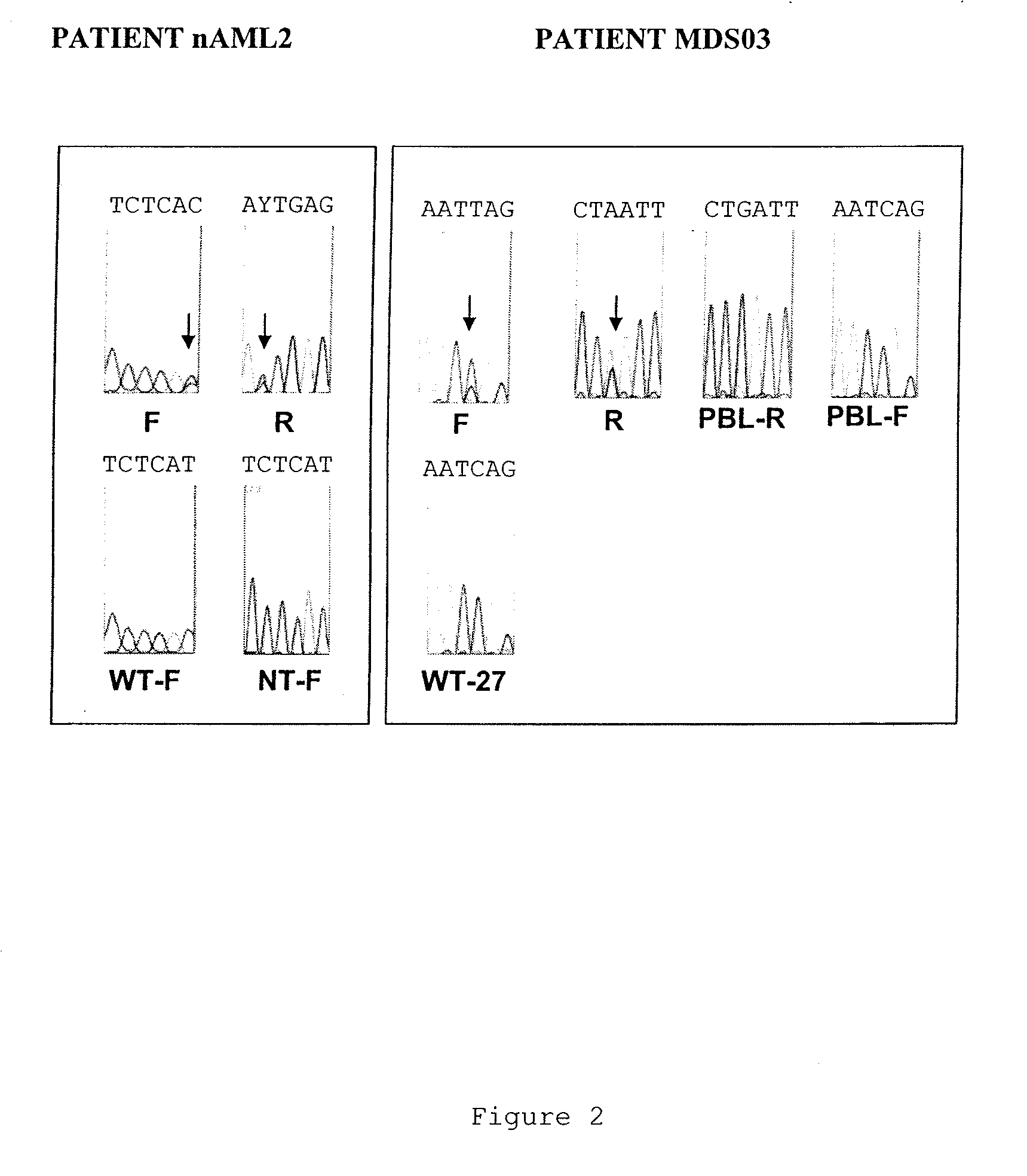
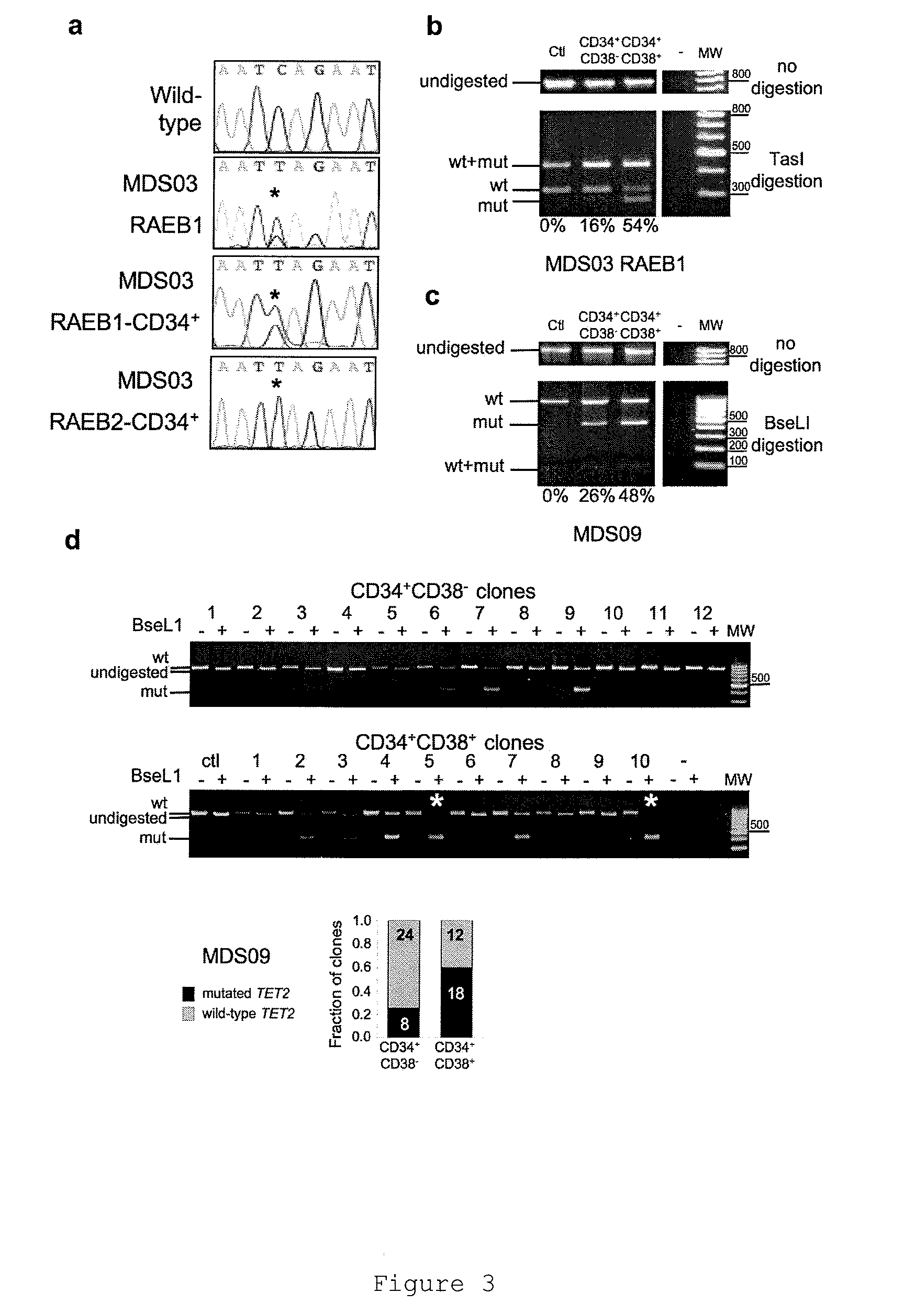
TET2 as a new diagnostic and pronostic marker in hematopoietic neoplasms
ActiveUS20110263523A1BiocideMicrobiological testing/measurementTen eleven translocationLymphatic neoplasm
The present invention concerns an in vitro method for diagnosing a myeloid tumour or a lymphoid tumour in a subject, which comprises the step of analyzing a biological sample from said subject by (i) detecting the presence of a mutation in the Ten Eleven Translocation protein family member 2 gene (TET2) coding for the polypeptide having the sequence SEQ ID NO:2, and / or (ii) analyzing the expression of the TET2 gene; wherein the detection of such a TET2 mutation, of the absence of expression of TET2 or of the expression of a truncated TET2 is indicative of a subject developing or predisposed to develop a myeloid tumour or a lymphoid tumour.
Owner:INST NAT DE LA SANTE & DE LA RECHERCHE MEDICALE (INSERM) +5
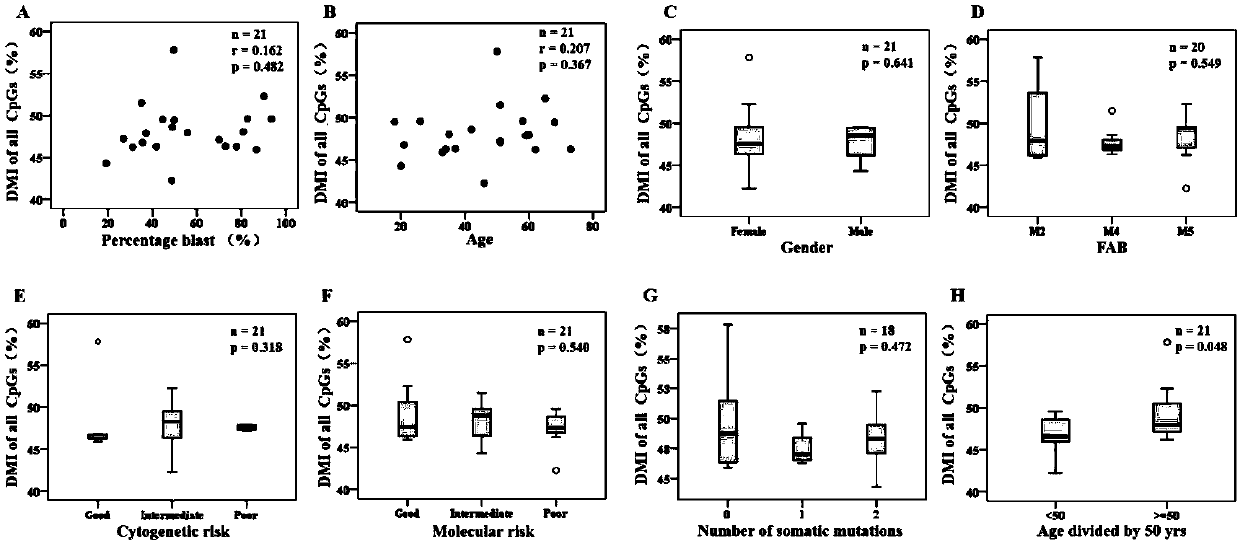
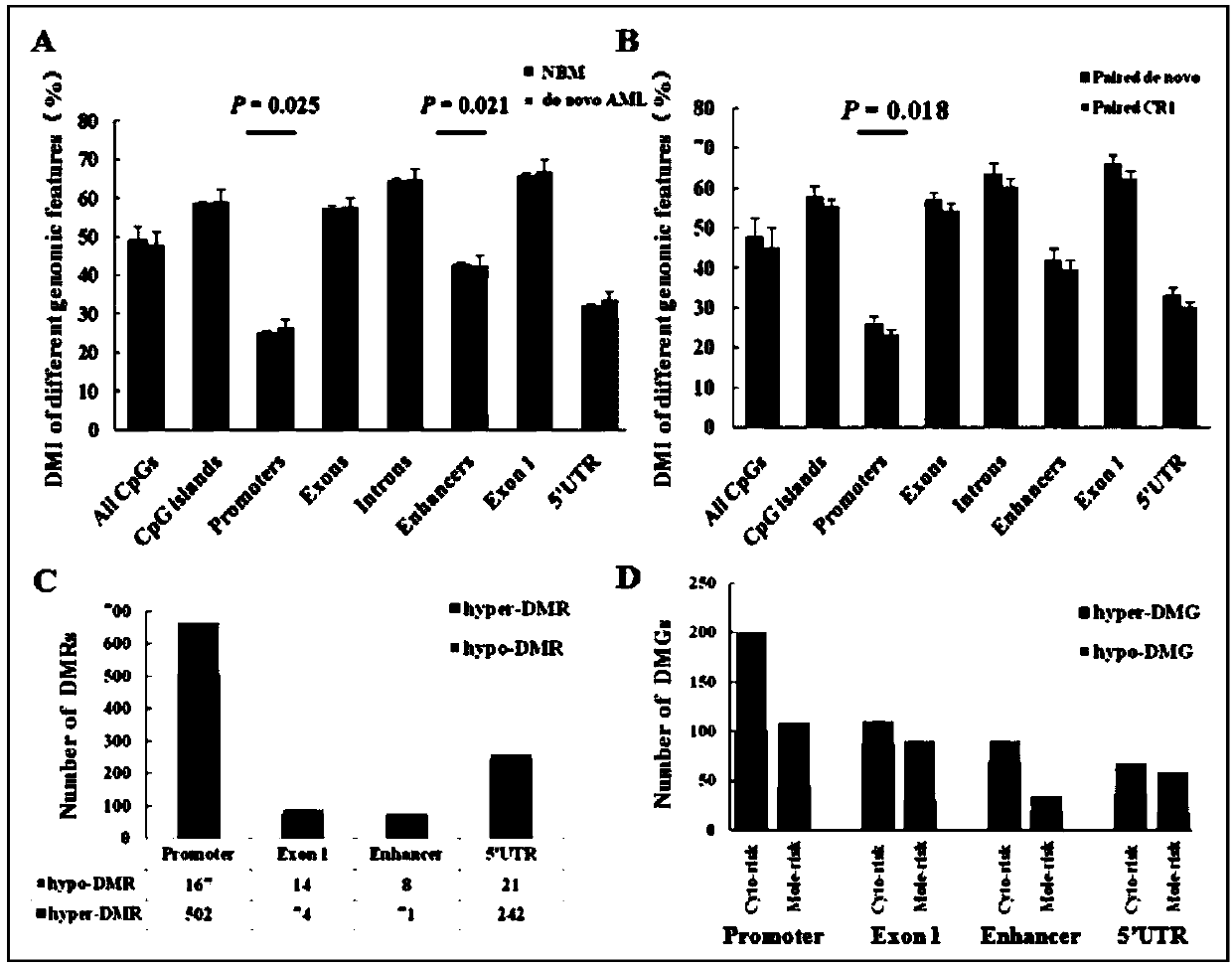
Method for screening prognostic markers of DNA methylation in acute myeloid leukemia
ActiveCN109852672AImproved prognosisIncrease conversion rateMicrobiological testing/measurementCpG sitePrognosis biomarker
The invention discloses a method for screening prognostic markers of DNA methylation in acute myeloid leukemia. The method includes the following steps: (1) sample and clinical data collection; (2) sample preparation; (3) genomic DNA extration; (4) whole-genome CpG site methylation capture sequencing; and (5)methylation sequencing data analysis and marker screening. The method uses the whole-genome CpG site methylation capture sequencing technology to detect DNA methylation in acute myeloid leukemia and perform prognosis analysis, and screens genetic markers which are regulated by DNA methylation and has prognostic significance through information of integrative genetics and epigenetics to guide accurate diagnosis and treatment of AML, thereby improving the curative effect and improving the prognosis of patients.
Owner:深圳豪石生物科技有限公司
Prognostic markers for prediction of treatment response and/or survival of breast cell proliferative disorder patients
InactiveUS20060024684A1Highly beneficialMicrobiological testing/measurementTreatment responseGenomic DNA
Aspects of the present invention provide compositions and methods for prognosis of, and / or predicting the estrogen treatment outcome of breast cell proliferative disorder patients, and in particular of patients with breast carcinoma. In preferred embodiments, this is achieved, at least in part, by determining the expression level of PITX2, and / or the genetic or the epigenetic modifications of the genomic DNA associated with the gene PITX2. Additional aspects of the invention provide novel sequences, oligomers (e.g., oligonucleotides or peptide nucleic acid (PNA)-oligomers), and antibodies, which have substantial utility in the described inventive methods and compositions.
Owner:EPIGENOMICS AG
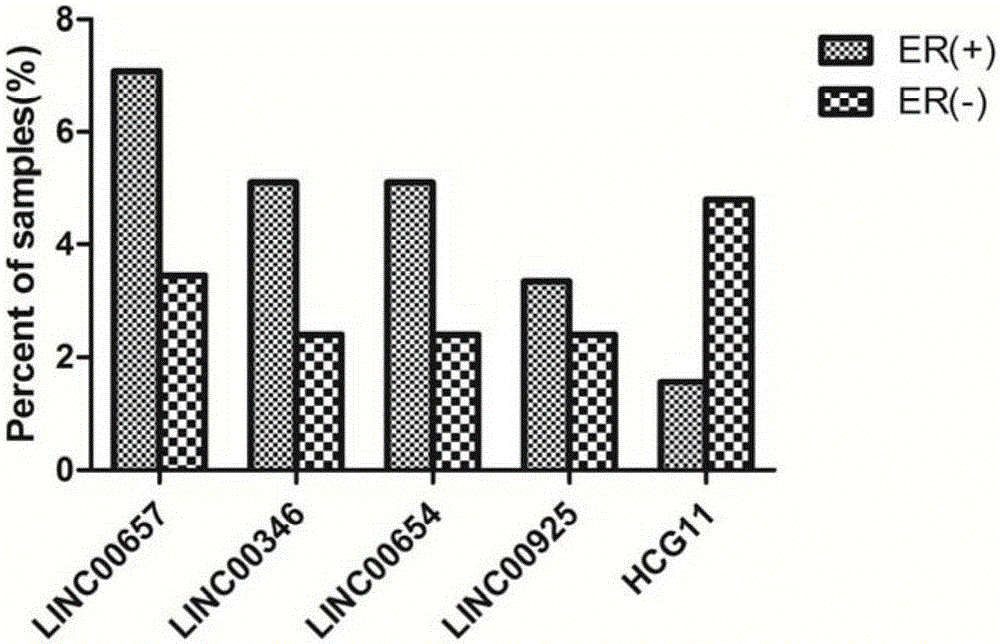
Breast tumor prognosis biomarker LncRNA detection method and clinical application thereof
InactiveCN105238782AWide detection rangeHigh sensitivityMicrobiological testing/measurementDNA/RNA fragmentationPrognosis biomarkerScreening method
The invention discloses a breast tumor prognosis biomarker LncRNA detection method and clinical application thereof, provides a group of new LncRNA in sequence shown as SEQ ID No1, SEQ ID No2, SEQ ID No3, SEQ ID No4 and SEQ ID No5 and relates to a method of taking specific LncRNA in samples (tissues, serum, urine, body fluid and the like) of patients suffering from breast tumors and corresponding juxtacancerous samples or normal samples as detection markers and biomarkers for prognostic auxiliary detection of breast tumors, related kits and a high-throughput sequencing and screening method. In-vitro diagnosis and judgment of canceration and process of breast tumors are realized by identification of differences of LncRNA expression quantity in the breast tumor samples and the corresponding juxtacancerous samples or normal samples. The invention further provides usage of the LncRNA as a breast tumor marker and probes and primers for LncRNA detection.
Owner:杭州壹锋生物科技有限公司 +1
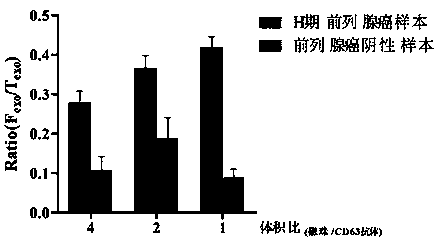
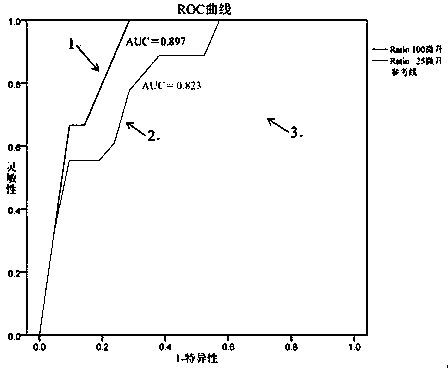
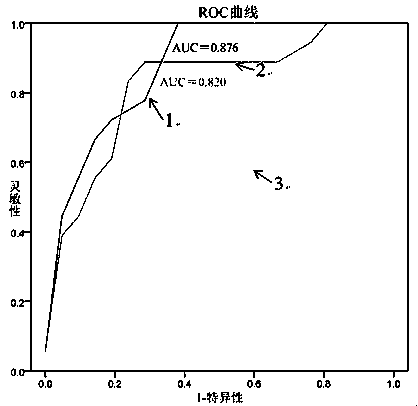
Prostatic cancer diagnosis, classification or prognosis marker, detection reagent or kit, system and application thereof
ActiveCN109507426AGood indicationHigh sensitivityBiological testingProstate cancerPrognosis prediction
The invention relates to a prostatic cancer diagnosis, classification and / or prognosis marker, a detection reagent or kit, a system and application thereof. The prostatic cancer diagnosis marker set disclosed by the invention is prepared from exosome-derived GCP2 and exosome-derived CD63 or GCP2 positive exosome and CD63 positive exosome. The prostatic cancer diagnosis marker set has the advantages of high flexibility and good specificity in prostatic cancer diagnosis, classification and / or prognosis prediction and has a good indication effect on prostatic cancer diagnosis, classification and / or prognosis prediction.
Owner:上海晟燃生物科技有限公司
Mitochondrial stress-70 protein markers for colorectal cancer
InactiveUS7700307B2Reduce and eliminate biological activityReduce expressionMicrobiological testing/measurementDisease diagnosisProtein markersATPase
Provided are previously uncharacterised markers of cancers, for example colorectal cancers, and uses of these as diagnostic and prognostic markers of cancers, and in particular colorectal cancers. The markers are SEQ ID NO: 1—hnRNP-K; SEQ ID NO:2—HMG-1; SEQ ID NO:3—proteasome subunit alpha type 1; SEQ ID NO:4—bifunctional purine biosynthesis protein; SEQ ID NO:5—ST11; SEQ ID NO:6—annex in IV; SEQ ID NO:7—60 kDa heat shock protein; SEQ ID NO:8—T complex protein 1 beta subunit; SEQ ID NO:9—T complex protein 1 epsilon subunit; SEQ ID NO: 10—mortalin; and SEQ ID NO: 11—TER-ATPase. The invention further provides related methods and materials for the use of the markers in therapeutic intervention in colorectal and other cancers e.g. to specifically target neoplastic cells without causing significant toxicity in healthy tissues, and to provide methods for the evaluation of the ability of candidate therapeutic compounds to modulate the biological activity of cancerous cells from the colon, rectum and other tissues.
Owner:AUVATION +2
Gene signatures for cancer diagnosis and prognosis
ActiveUS20130302242A1In-vivo radioactive preparationsNucleotide librariesPrognosis biomarkerCancers diagnosis
Owner:MYRIAD GENETICS
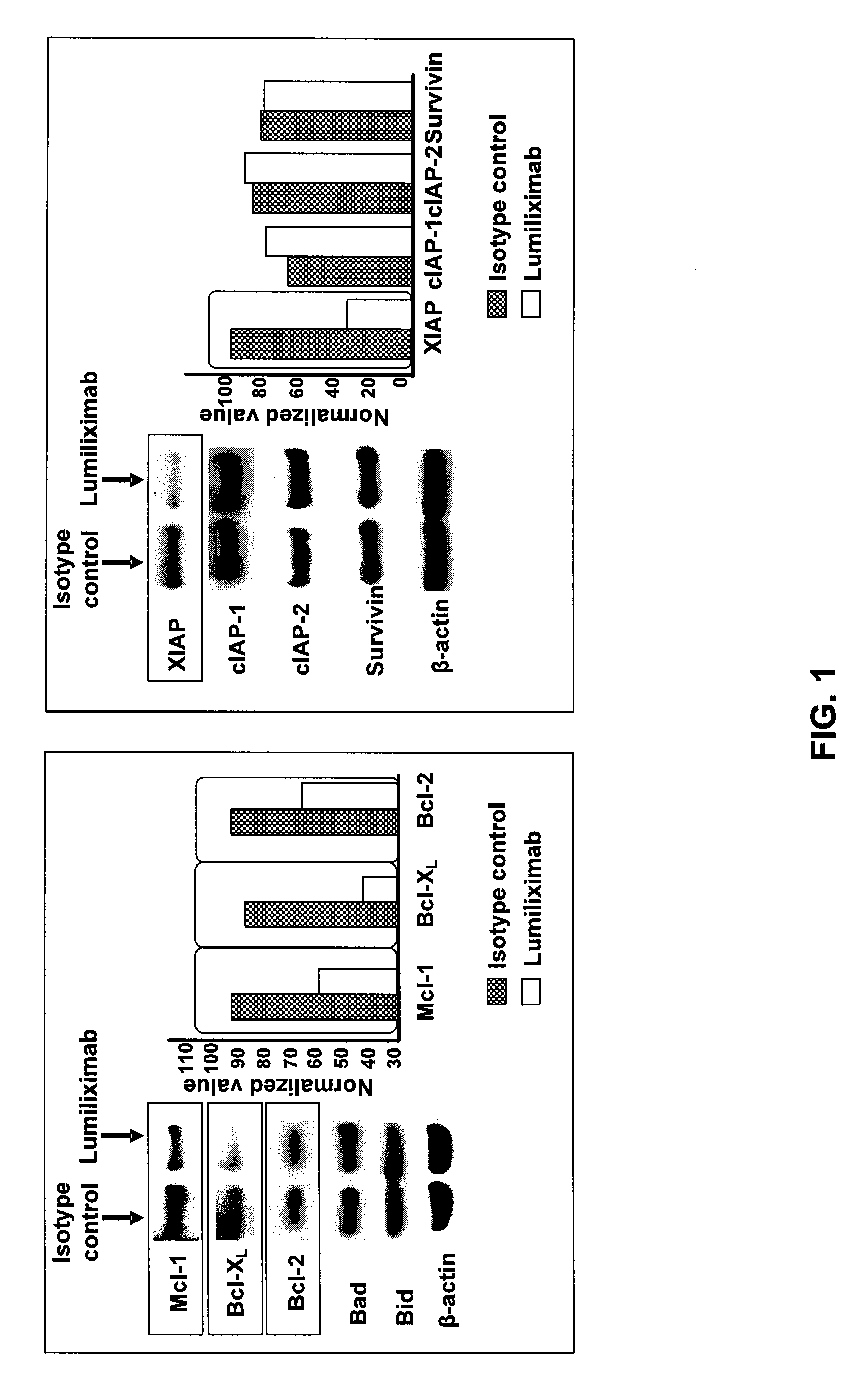
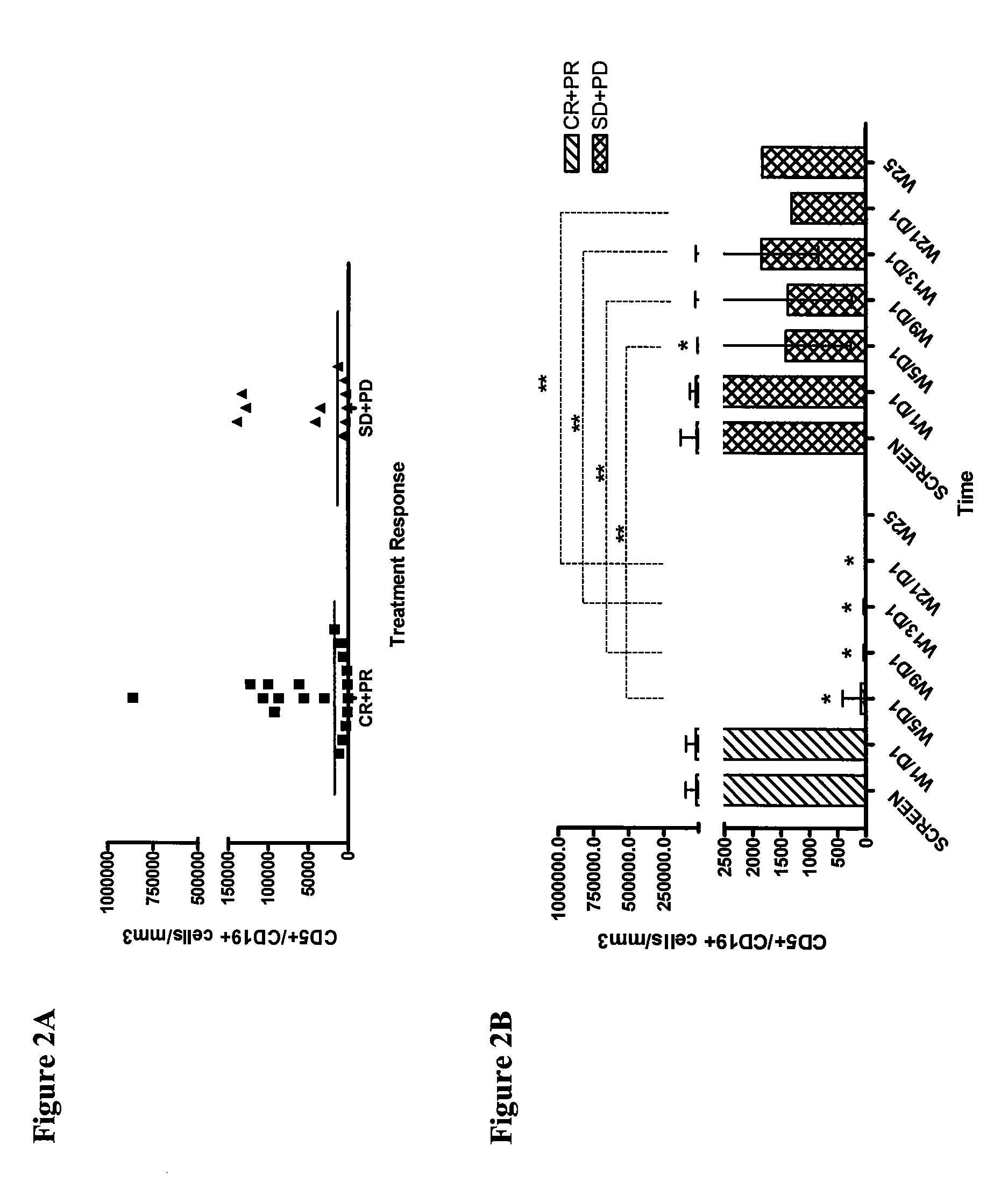
Use of CD23 Antibodies to Treat Malignancies in Patients with Poor Prognosis
InactiveUS20090252725A1Induce apoptosisTherapeutically effectiveAntibody ingredientsTissue cultureMammalZAP70
The invention relates to methods of treating B-cell chronic lymphocytic leukemia, and other CD23+ malignancies, in patients with poor prognostic markers. The method comprises administration of an CD23 antibody, including for example, lumiliximab to a mammal that over expresses a poor prognostic marker. The method can also comprise administration of lumiliximab in combination with fludarabine, cyclophosphamide and rituximab. Patients with poor prognostic markers include, for example, patients that overexpress ZAP70, CD38, β2-microglobulin and / or soluble CD23.
Owner:BIOGEN IDEC MA INC
Evidence-based method for screening gastric cancer prognosis molecular markers
InactiveCN103310129ASignificant prognostic indicatorsClear sourceSpecial data processing applicationsScreening durationScreening method
The invention discloses an evidence-based method for screening gastric cancer prognosis molecular markers. The method comprises the steps as follows: 1), an inclusion criterion is set for a gastric cancer prognosis biomarker experimental research; 2), a research evidence retrieval strategy is set; 3), the prognosis biomarkers are screened preliminarily; 4), research data are extracted; 5), quality assessment on the experimental research quality is performed; 6), standardized data are processed; 7), the prognosis biomarkers are screened; and 8), according to the screening criterion, the gastric cancer prognosis biomarkers having potential application values are screened, and the prognosis values are described. In conclusion, the evidence-based method for screening the gastric cancer prognosis biomarkers is provided, and the gastric cancer prognosis biomarkers are screened, so that the screening cost is reduced, the screening time is shortened, professional thresholds are reduced, and the application values of the prognosis biomarkers are increased.
Owner:ZHEJIANG UNIV
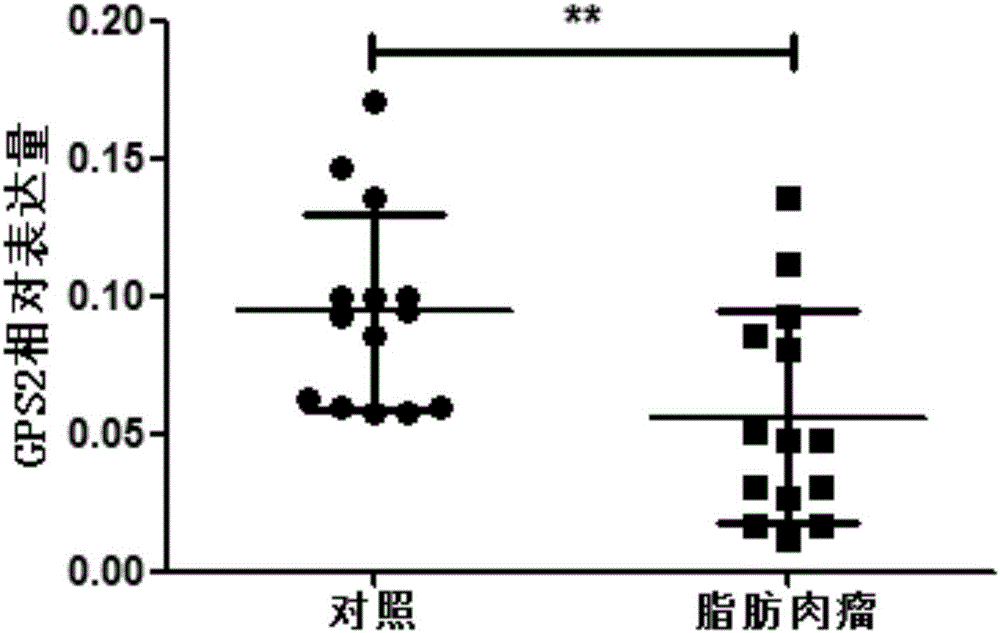
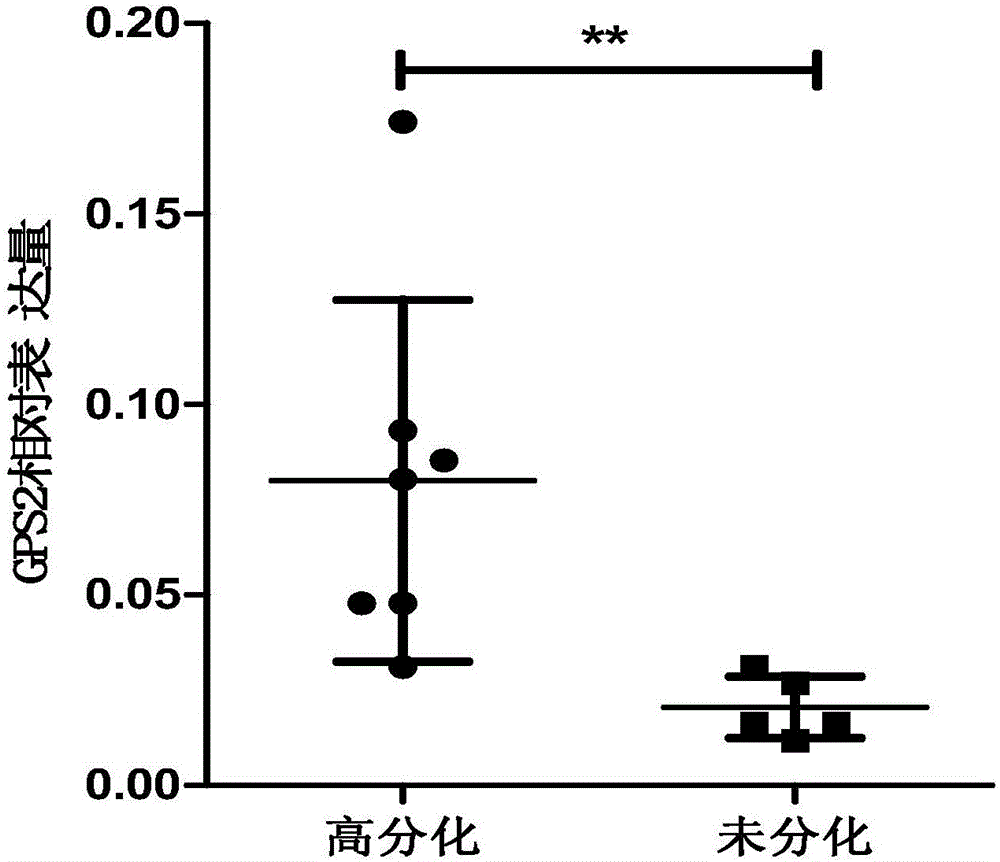
Application of GPS2 gene in preparation of drugs for prognosis, diagnosis or prevention and treatment of soft tissue tumor
ActiveCN106075468APeptide/protein ingredientsMicrobiological testing/measurementAbnormal tissue growthDisease
The invention belongs to the field of biological medicines and relates to an application of a GPS2 gene in preparation of drugs for prognosis, diagnosis or prevention and treatment of soft tissue tumor. The GPS2 gene is in low expression in soft-tissue sarcoma such as liposarcoma, and proliferation of the liposarcoma can be promoted by inhibiting the expression of the GPS2 gene. The expression of the GPS2 gene has obvious relevance with prognosis of the liposarcoma, which shows that the GPS2 gene can be used for clinical pathological diagnosis of patients with liposarcoma. The GPS2 gene can be used as a tumor-suppressing gene in the liposarcoma, and can be used as a possible prognostic marker and a therapeutic molecule of the liposarcoma.
Owner:INST OF RADIATION MEDICINE ACAD OF MILITARY MEDICAL SCI OF THE PLA
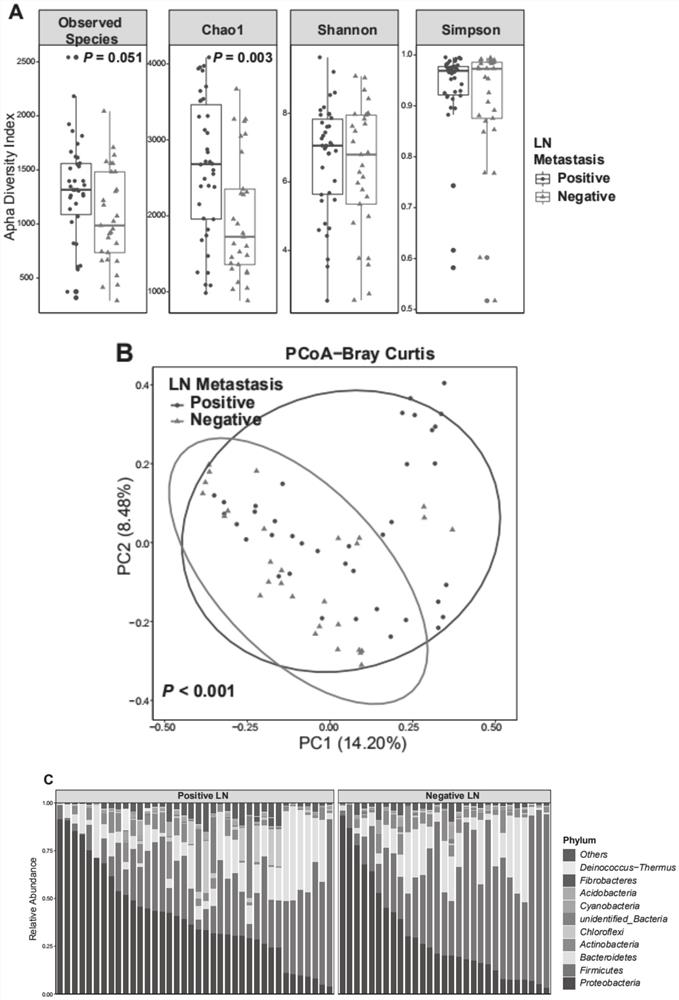
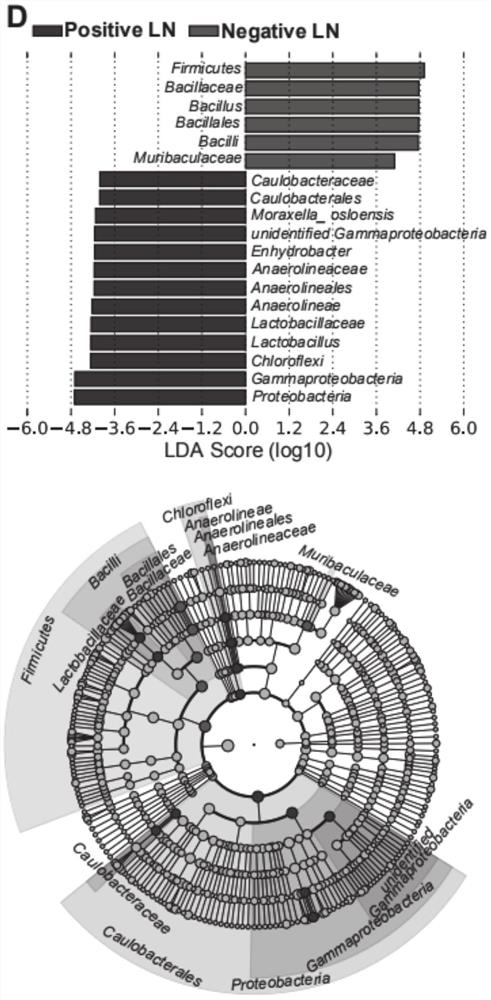
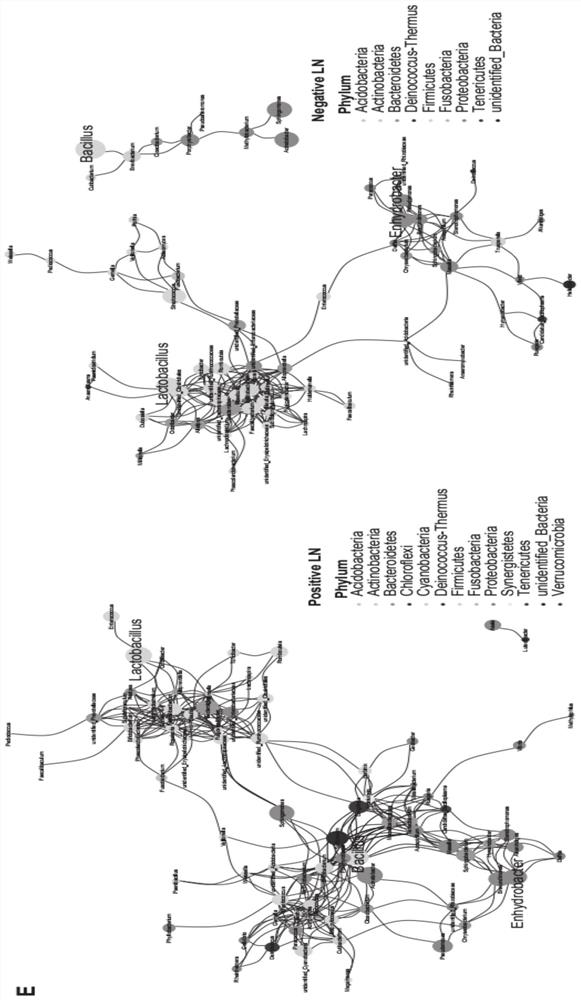
HNC (Head and Neck Cancer) prognosis biomarker based on lymph node microbial flora and application of HNC prognosis biomarker
PendingCN113684242AGood forecastThe value of good practical applicationHealth-index calculationMicrobiological testing/measurementFloraMicrobiome
The invention provides an HNC (Head and Neck Cancer) prognosis biomarker based on a lymph node microbial flora and application of the HNC prognosis biomarker, and belongs to the technical field of disease prognosis and molecular biology. According to the invention, the 16S ribosomal RNA gene of the lymph node microbial flora of an HNC patient is subjected to high-throughput sequencing, so that it is proved that different species exist in flora species of metastatic lymph nodes and non-metastatic lymph nodes in the HNC patient; furthermore, through a microbial flora symbiotic relationship network, it is found that the flora interrelation between the species of the metastatic lymph node and the species of the non-metastatic lymph node of the HNC patient is also different at the genus level; the result discovers the characteristics of the microbiome of the metastatic lymph node and the non-metastatic lymph node of the HNC patient for the first time; and meanwhile, it is known by analysis that the flora difference characteristics (diversity index values and relative abundance of different species) have a good prediction effect on the lifetime (total lifetime and three-year lifetime) of the HNC patient, so that the invention has a good practical application value.
Owner:SHANDONG UNIV +1
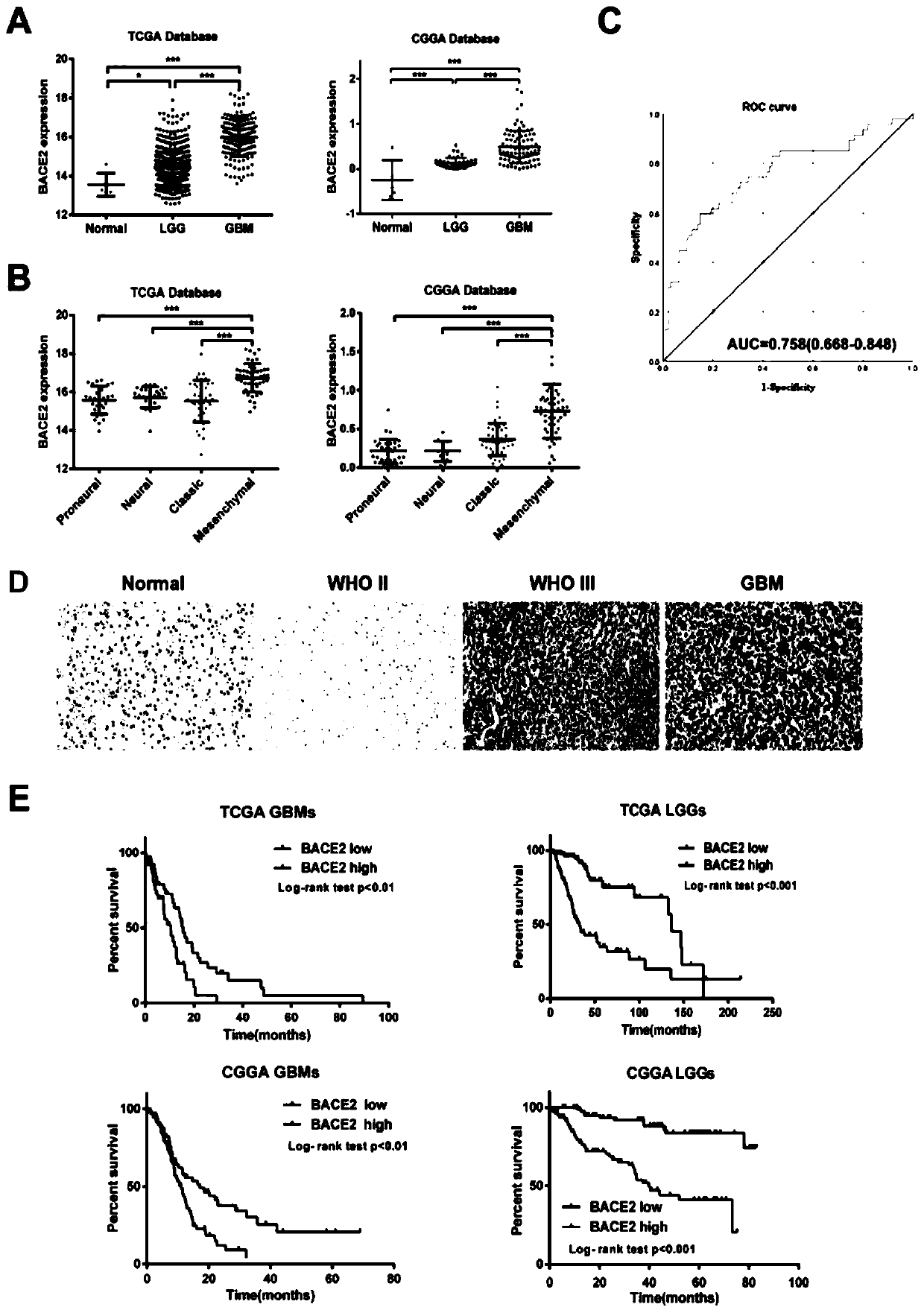
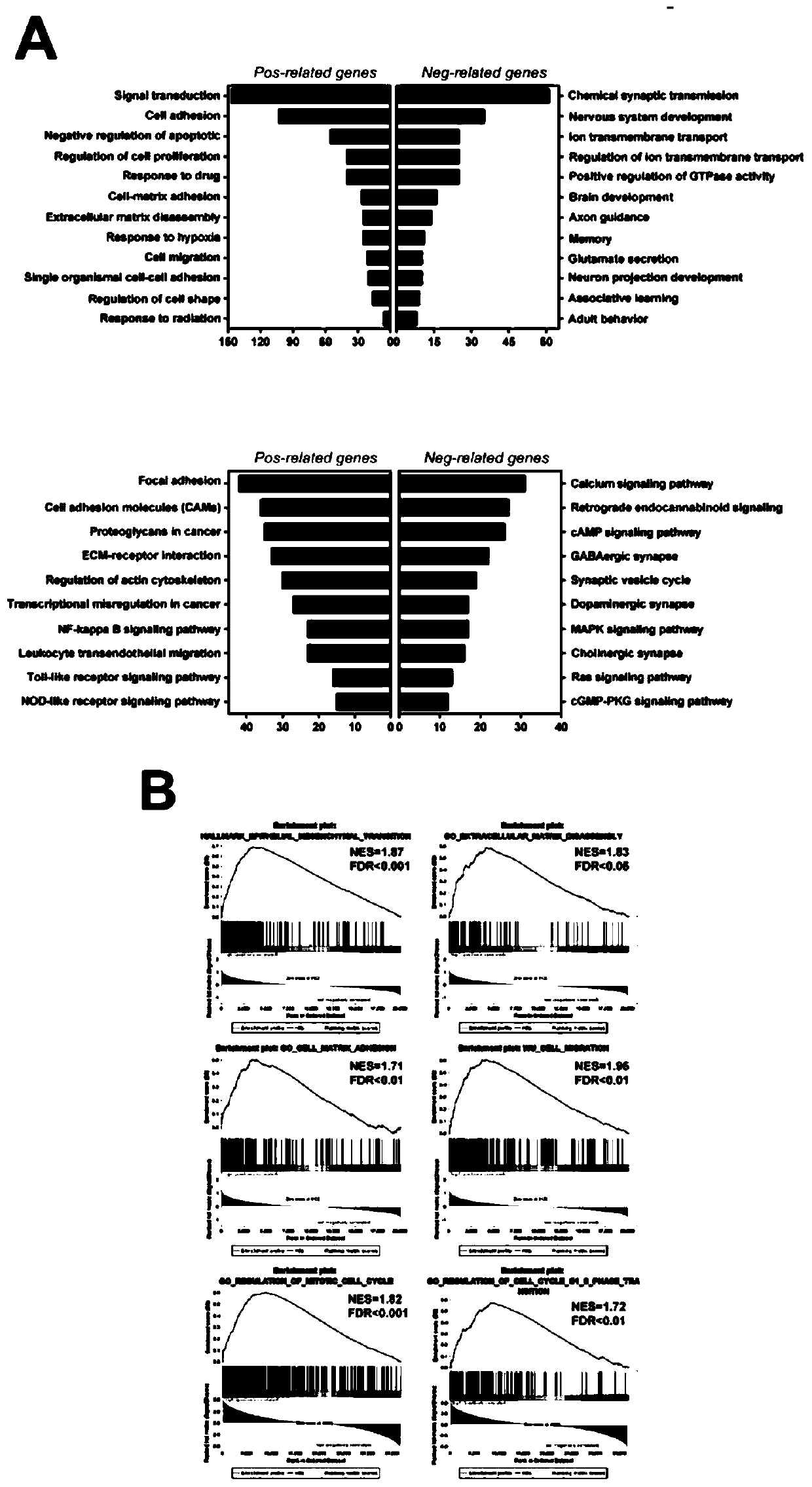
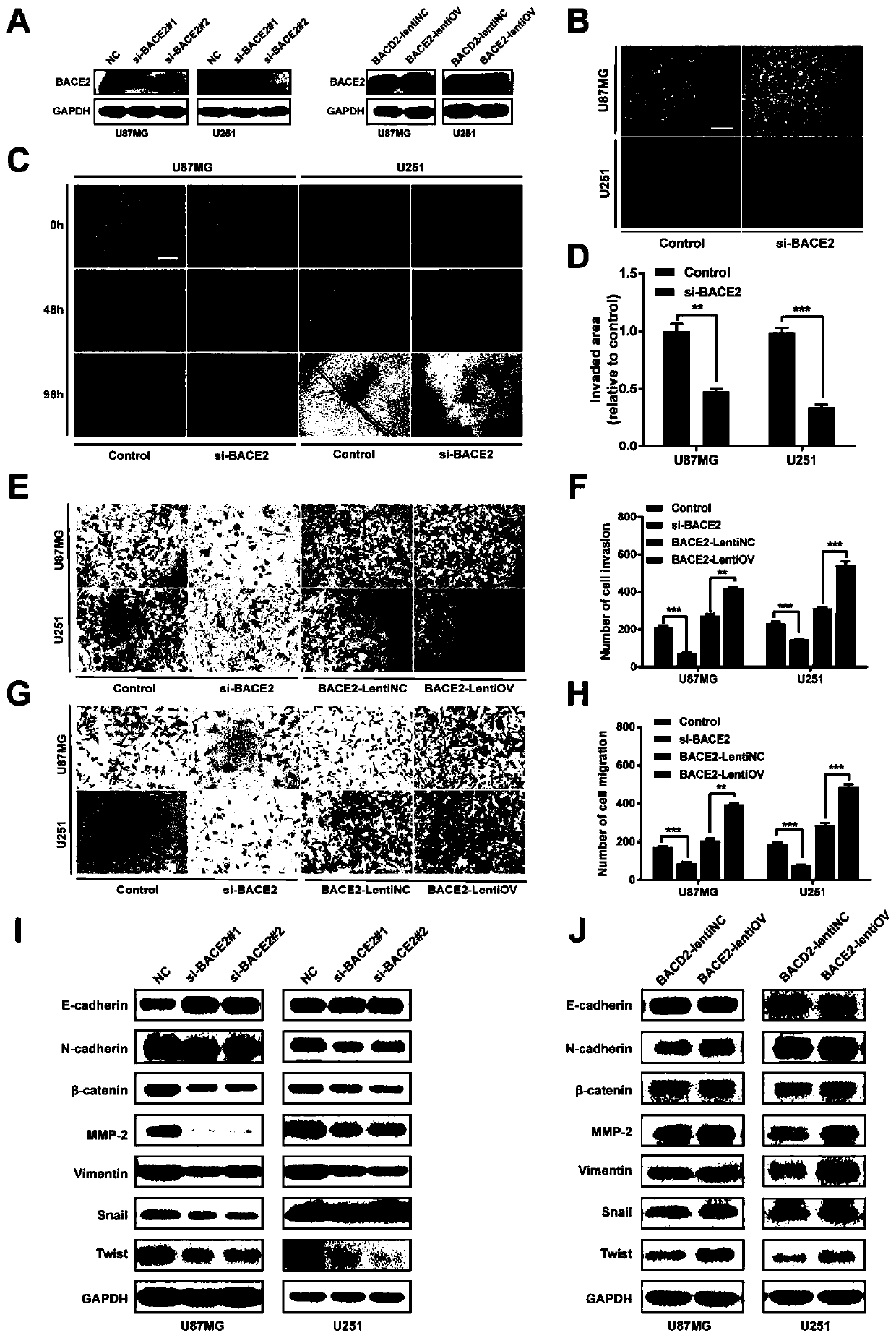
Application of BACE2 as glioma prognosis/diagnosis/treatment marker
ActiveCN110747276APromote invasionEasy transferMicrobiological testing/measurementBiological material analysisSMADPrognosis biomarker
The invention belongs to the technical field of glioma prognosis markers, and particularly relates to application of BACE2 as a glioma prognosis / treatment marker. According to the invention, the research results prove that high-level BACE2 expression is related to the high-level human brain glioma, the mesenchymal molecule subtype of GBM and the poor prognosis for the first time; furthermore, TGF[beta] 1 stimulates BACE2 expression by means of Smad dependent signal transduction, so as to regulate the activity of TNF-[alpha] induced NF-[kappa] B by means of a PP1 / IKK pathway to promote tumorigenesis in glioma cells. The BACE2 can be used as a novel biomarker and a potential treatment target for treating human brain glioma.
Owner:SHANDONG UNIV QILU HOSPITAL
Cytokines as prognostic markers of respiratory-tract infection following major surgery
InactiveUS20120129176A1Reduces and excludes variationHigh riskHealth-index calculationMicrobiological testing/measurementFactor iiPre operative
The invention relates to the use of a certain subset of cytokine markers as prognostic variables of infection status in an individual, and especially as prognostic markers of a patients developing severe infection such as pneumonia, and respiratory tract infection following surgery. The subset of cytokine markers consists of the interleukin cytokines IL-2, IL-7, IL-23, IL-27, and IL-IO, and Interferon-γ (INFγ) and Tissue Necrosis Factor-α (TNFα). The markers may be employed as individual prognostic variables of infection status, or they may be used in pairs or other combinations. Generally, the abundance of the markers is correlated with infection status by means of an absolute pre-operative value of biomarker abundance, ratio's of pre-operative to post-operative biomarker abundance, or ratio values for pairs of certain biomarkers within the subset. Typically, cytokine abundance is expressed in terms of mRNA copy number wherein the copy numbers are ideally normalised to a house keeping gene and quantification of mRNA copy number is determined by RT-PCR containing reference serial dilutions of cytokine specific cDNA.
Owner:THE PROVOST FELLOWS & SCHOLARS OF THE COLLEGE
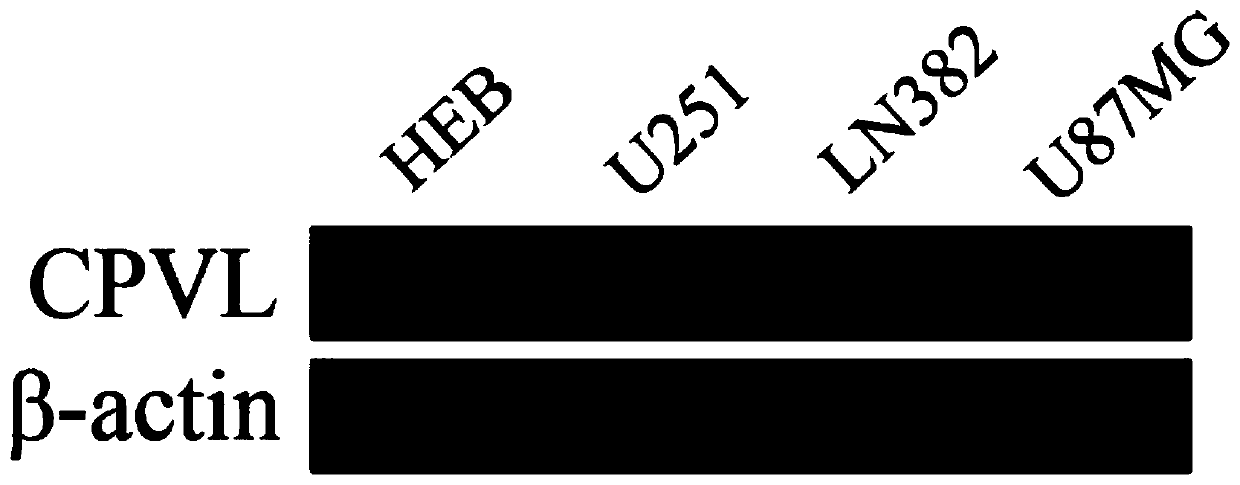
Glioma prognosis marker CPVL and application thereof
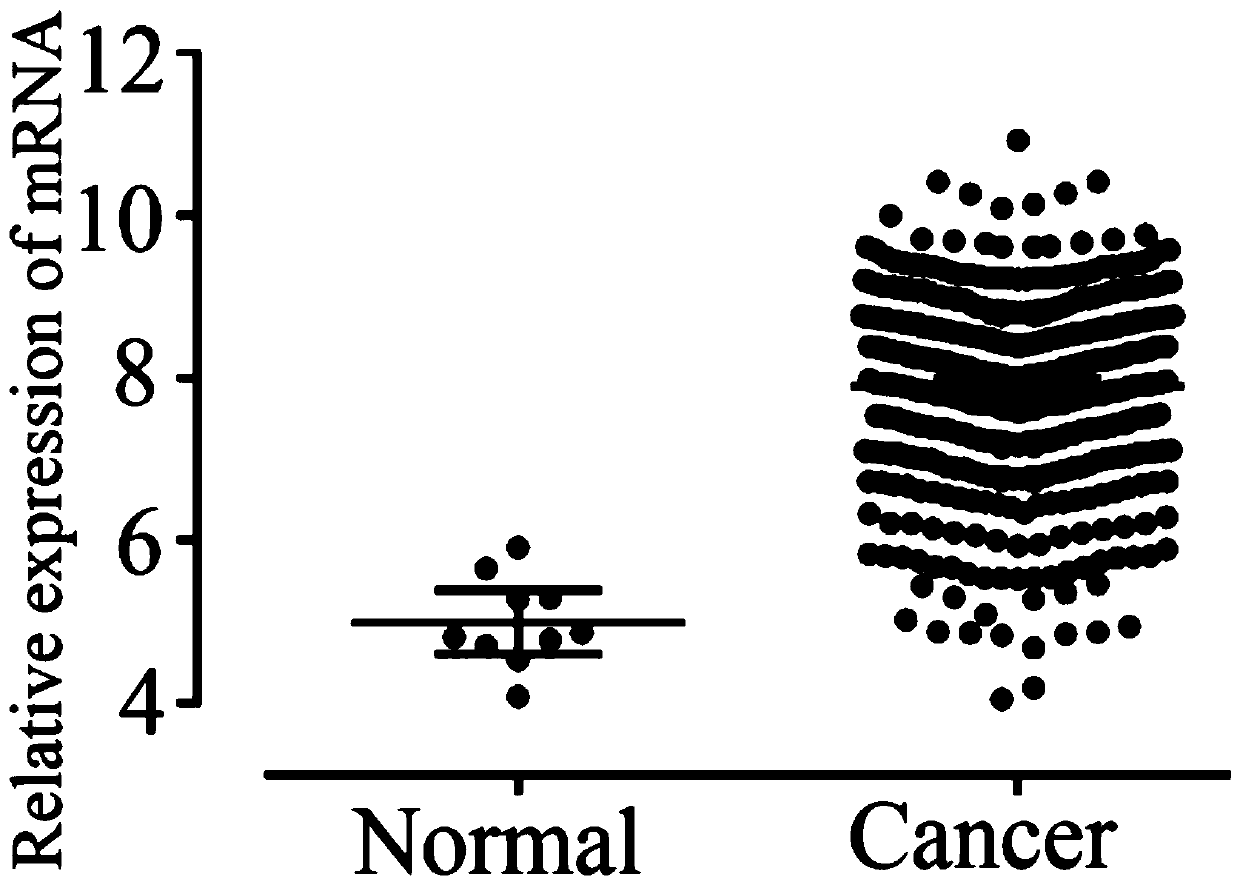
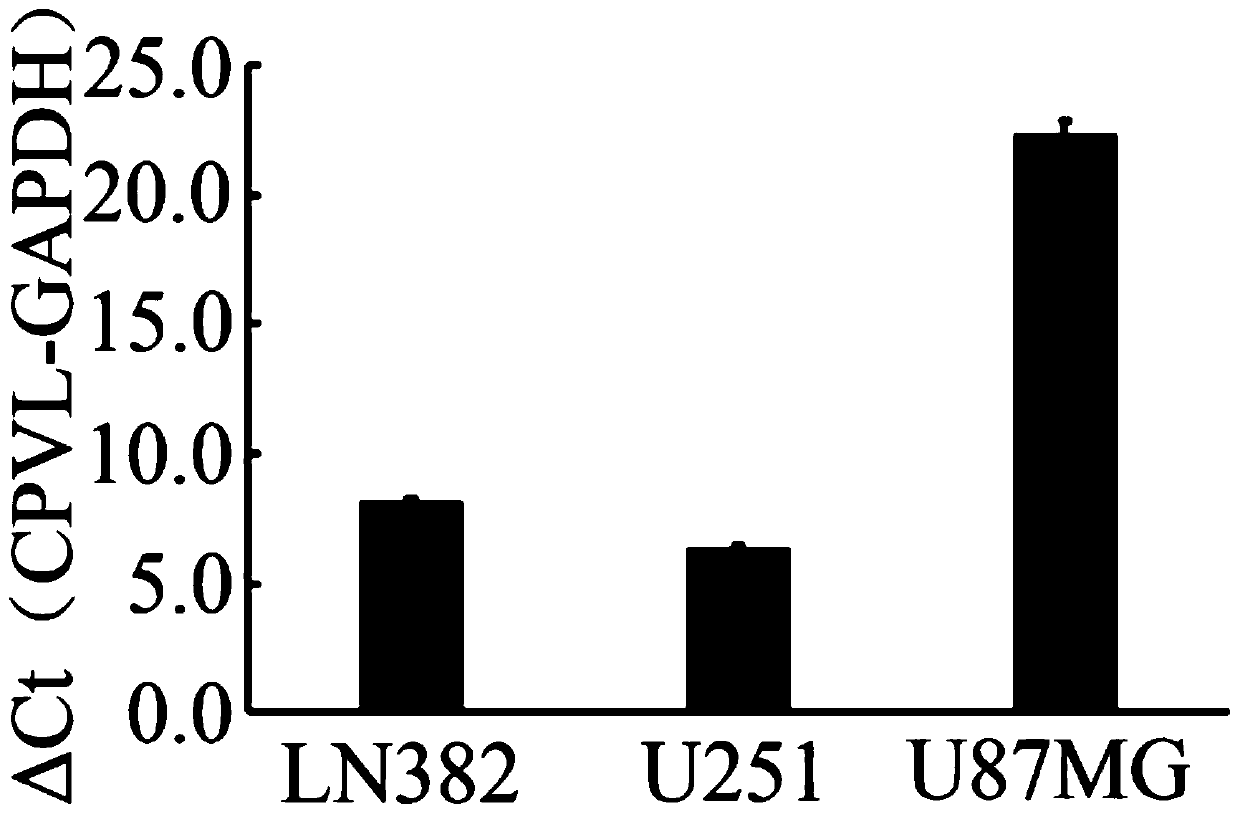
ActiveCN109825587APromote apoptosisInduce apoptosisMicrobiological testing/measurementDNA/RNA fragmentationBiologyTreatment targets
The invention relates to the technical field of biology, in particular to a glioma prognosis marker CPVL and an application thereof. Thetechnical scheme includes: a glioma prognosis marker CPVLmRNA, the application of a reagent for detecting the expression amount of the marker CPVLmRNA in glioma tissues in preparing a glioma prognosis preparation; a glioma prognosis kit capable of detecting the content of the marker CPVLmRNA in glioma tissues, comprising PCR primers for detecting the content of the marker CPVLmRNA; and application of a glioma prognosis marker CPVLmRNA. The glioma prognosis marker CPVL and the application thereof can effectively verify that the CPVL knock-down can promote the apoptosis of glioma cells, inhibit the proliferation and the tumorigenesis of the glioma cells, regulate the cell cycle of the glioma, induce the apoptosis of the glioma cells by the CPVL knock-down, clarify the pathogenesis of the glioma and have important theoretical and practical meanings, and the oncogene CPVL plays an important role in the generation and development of the glioma, thereby providing a potential treatment target point for the treatment of the glioma.
Owner:THE FIRST AFFILIATED HOSPITAL OF WANNAN MEDICAL COLLEGE YIJISHAN HOSPITAL OF WANNAN MEDICAL COLLEGE
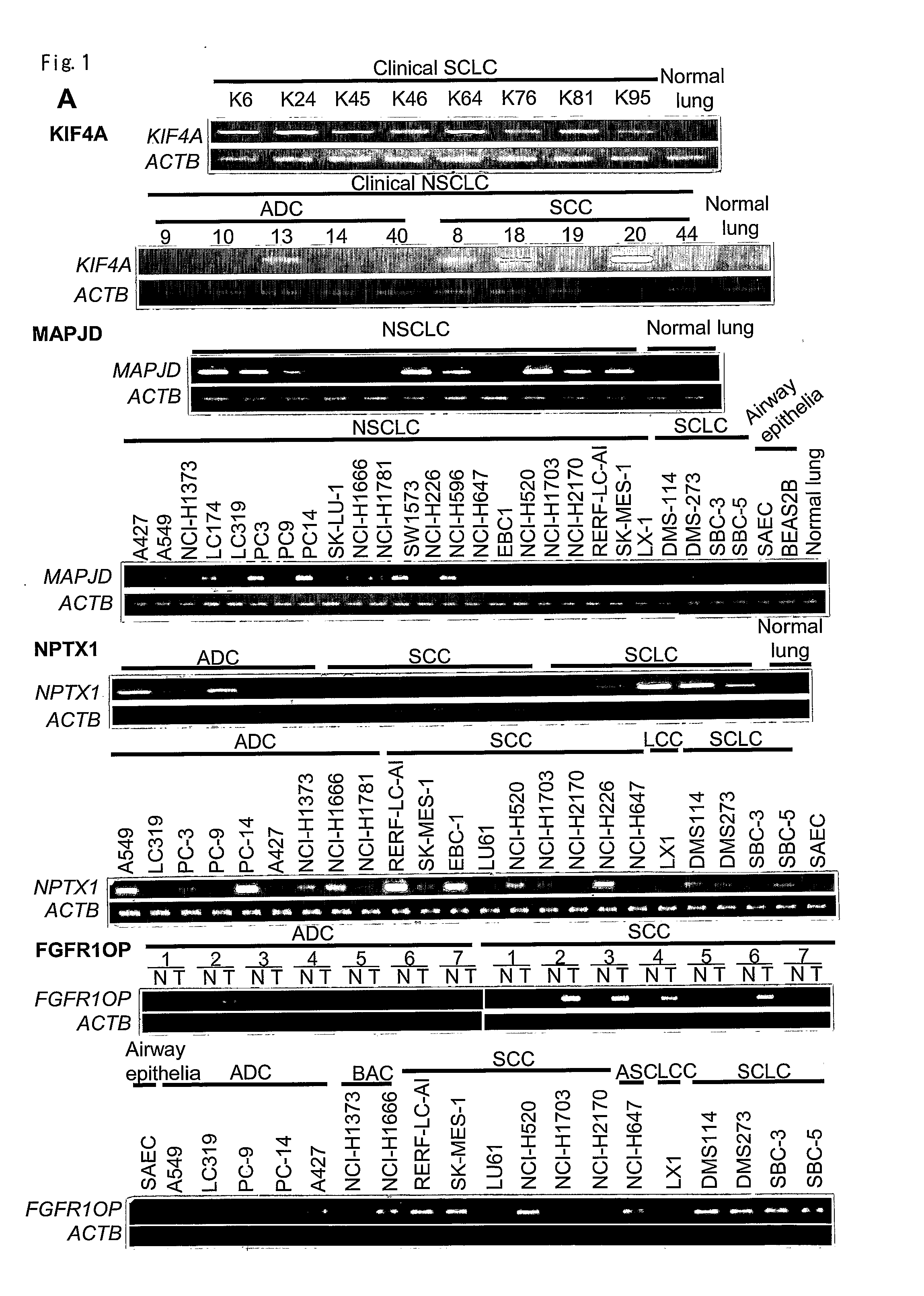
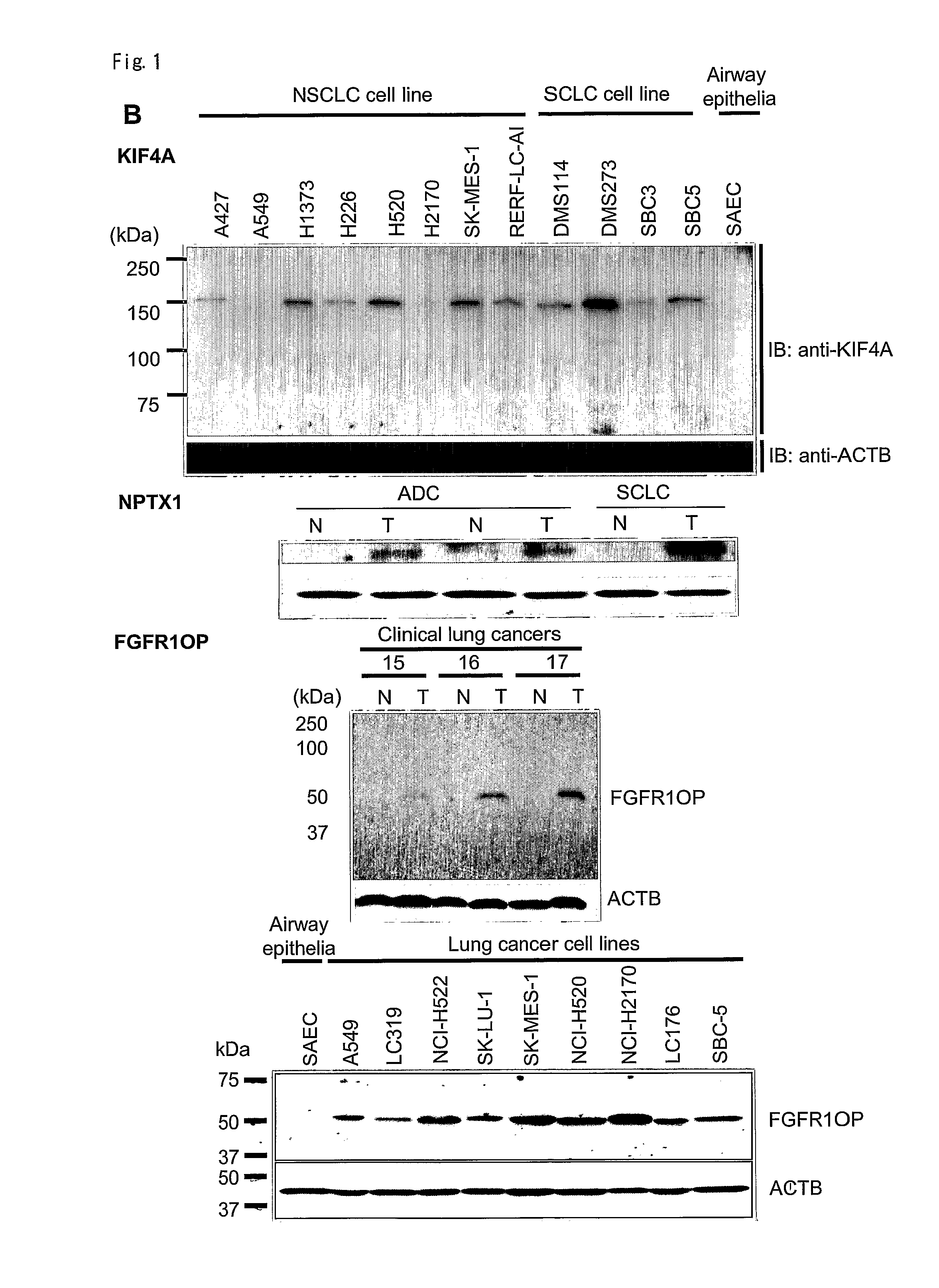
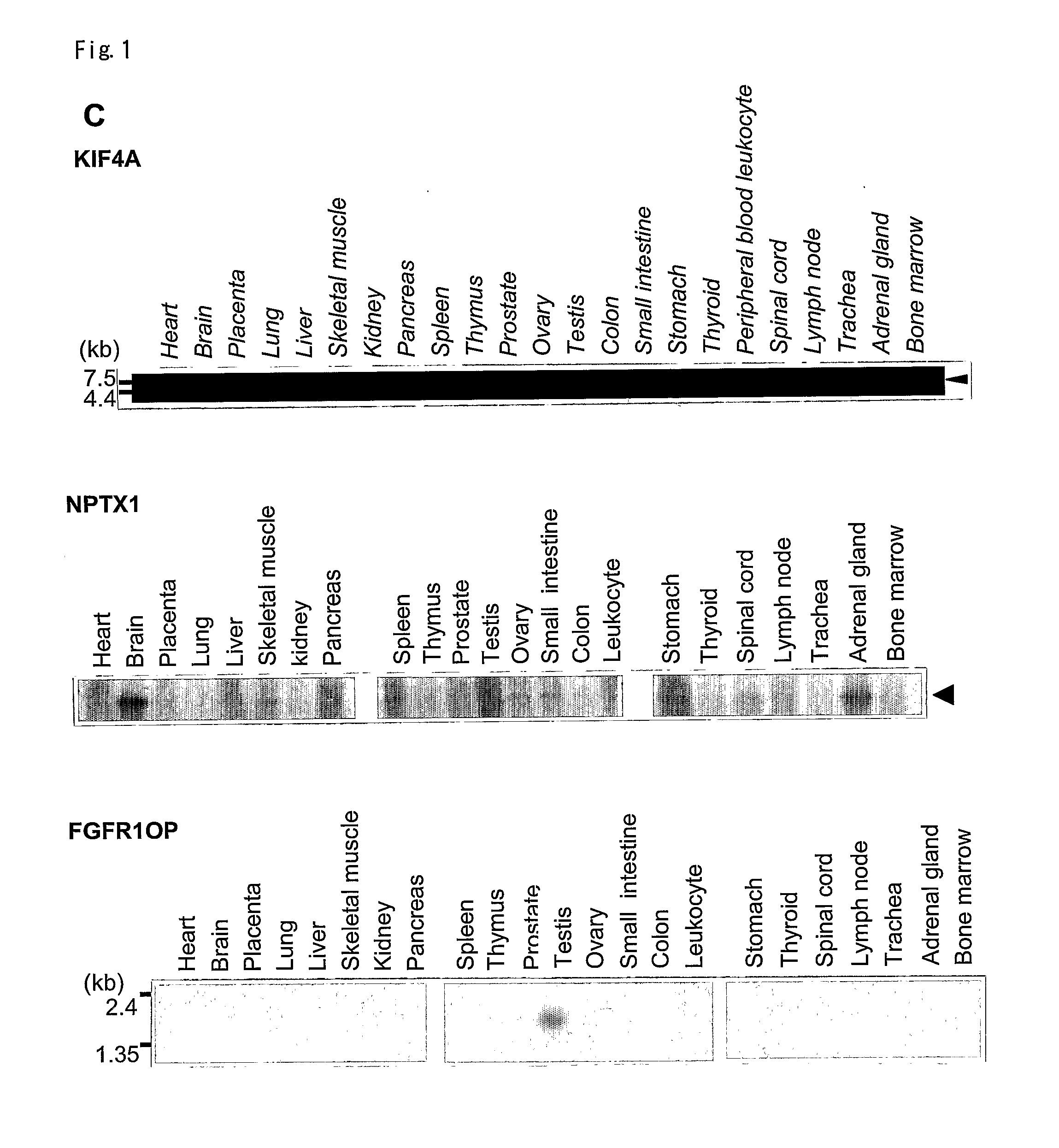
Prognostic markers and therapeutic targets for lung cancer
InactiveUS20100292090A1Easy to useHigh expressionMicrobiological testing/measurementLibrary screeningLung cancerCancer research
The present invention provides a diagnostic marker to detecting lung cancer. In particular, the present invention provides lung cancer marker genes i.e. KIF4A, MAPJD, NPTX, or FGFR1OP. The present invention further provides methods and kit for identifying compounds for treating lung cancer as well as methods for predicting a prognosis or diagnosis of lung cancer. In particular, the present invention provides methods and kits for identifying inhibitors of the interaction between KIF4A / ZNF549, KIF4A / ZNF553, MAPJD / MYC, or FGFR1OP / WRNIP1 which find utility in the treatment and prevention of lung. Alternatively, the present invention provides MAPJD associated with HAT complex as therapeutic target.
Owner:ONCOTHERAPY SCI INC
Cytokines as prognostic markers of respiratory-tract infection following major surgery
InactiveUS20150197808A1Reduce riskHigh riskHealth-index calculationMicrobiological testing/measurementInterferon alphaPre operative
The invention relates to the use of a certain subset of cytokine markers as prognostic variables of infection status in an individual, and especially as prognostic markers of a patients developing severe infection such as pneumonia, and respiratory tract infection following surgery. The subset of cytokine markers consists of the interleukin cytokines IL-2, IL-7, IL-23, IL-27, and IL-10, and Interferon-γ (INFγ) and Tissue Necrosis Factor-α (TNFα). The markers may be employed as individual prognostic variables of infection status, or they may be used in pairs or other combinations. Generally, the abundance of the markers is correlated with infection status by means of an absolute pre-operative value of biomarker abundance, ratio's of pre-operative to post-operative biomarker abundance, or ratio values for pairs of certain biomarkers within the subset. Typically, cytokine abundance is expressed in terms of mRNA copy number wherein the copy numbers are ideally normalised to a house keeping gene and quantification of mRNA copy number is determined by RT-PCR containing reference serial dilutions of cytokine specific cDNA.
Owner:TRINITY COLLEGE DUBLIN
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com