Nucleic acid expanding gene chip hybridization intellectualization and inspecting instrument thereof
A technique for amplifying genes and chip hybridization, which is applied in biochemical equipment and methods, and microbial measurement/inspection. PCR time and the effect of improving work efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
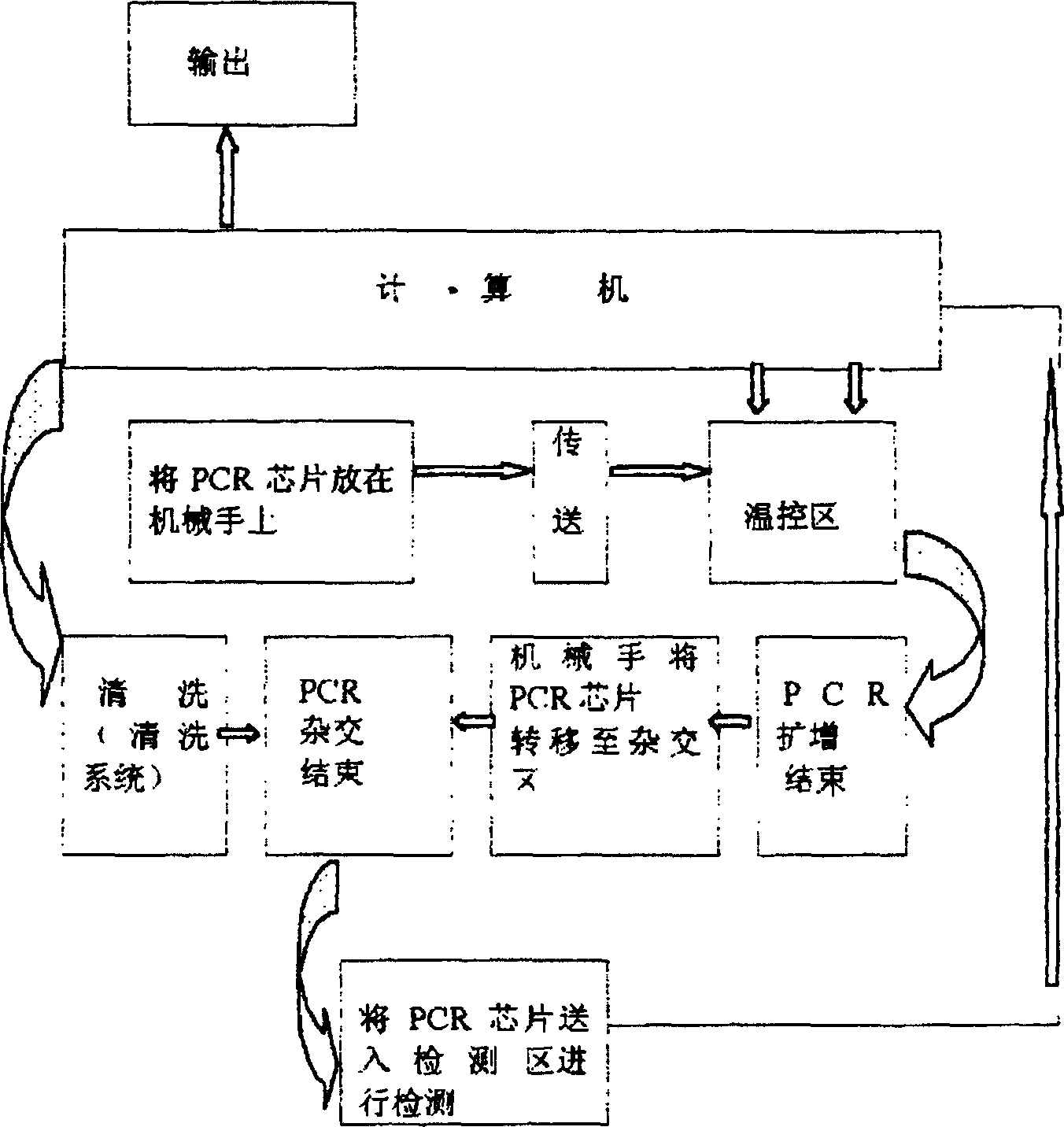
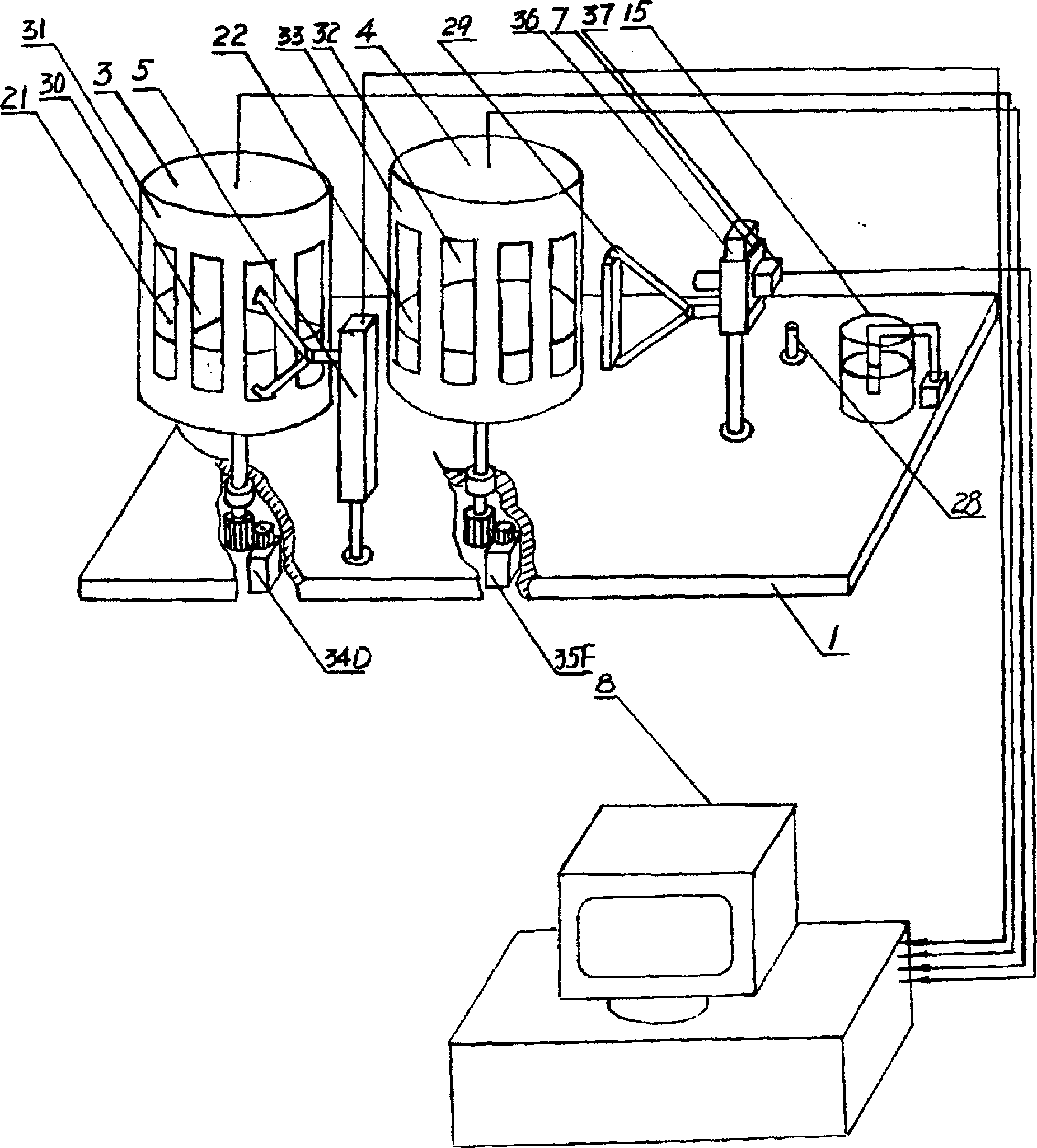
[0032] As shown in the figure, the nucleic acid amplification gene chip hybridization intelligent detection method is characterized by the following steps:
[0033] (A) Send the PCR chip into the temperature control area, and complete the denaturation, annealing, and extension processes of the PCR process according to the set temperature mode;
[0034] (B) After the PCR cycling process is completed, move the PCR chip to the hybridization area, and control the temperature of the entire hybridization process according to the set temperature:
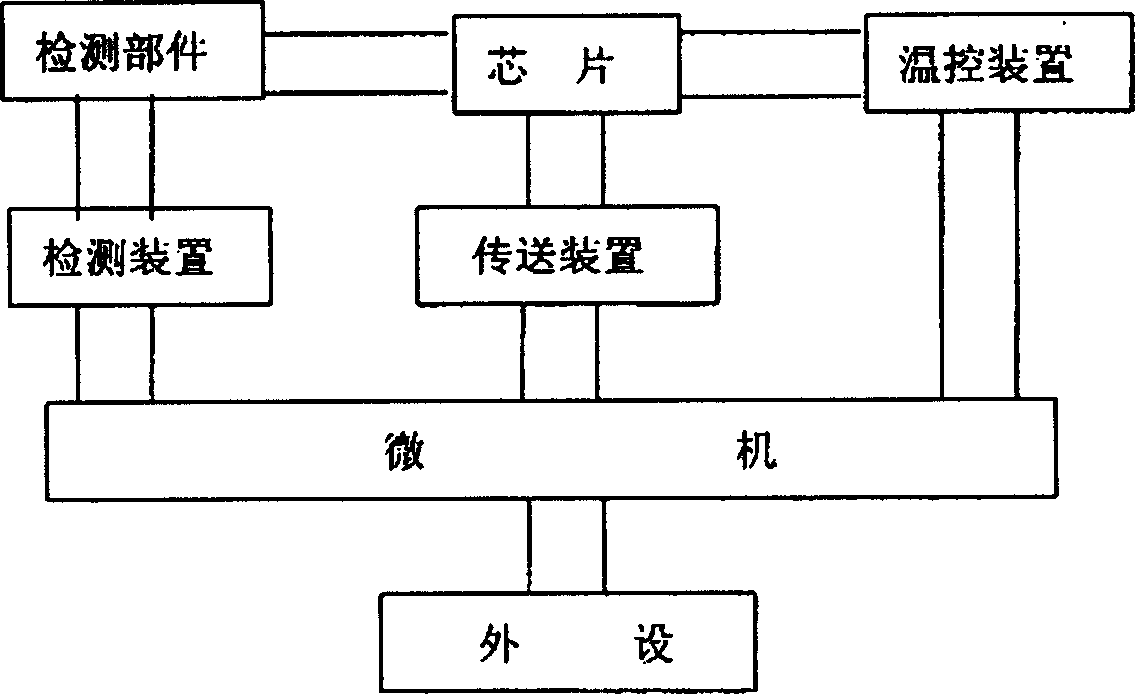
[0035] (C) After the hybridization process is completed, the PCR chip is cleaned by the cleaning device. After the cleaning is completed, the gene chip is sent into the detection area by the transmission device for detection. The position signal of the detection device and the position signal of the hybridization point of the PCR chip are determined by the sensor, The photoelectric converter is output to the computer, the analog signal is ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com