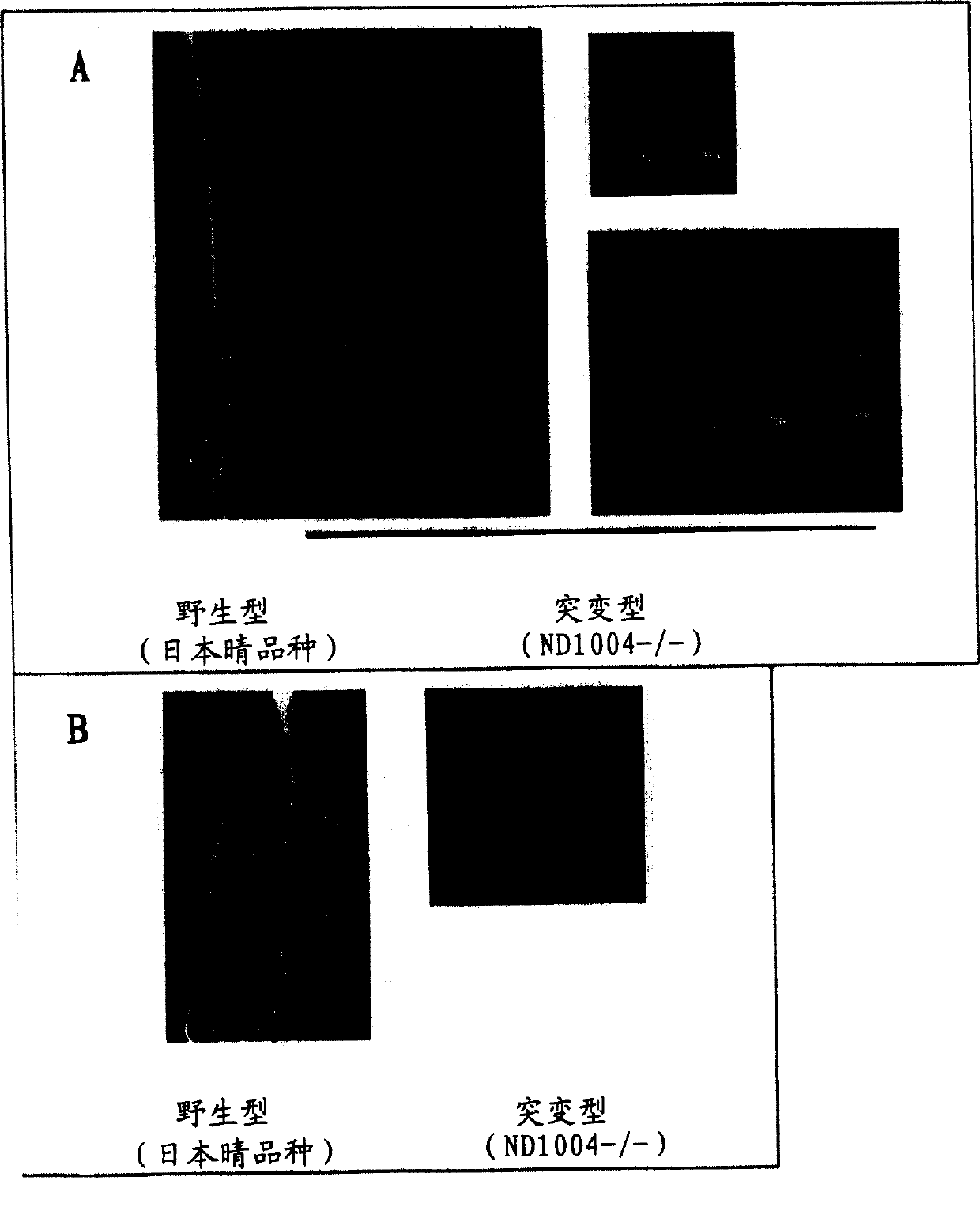
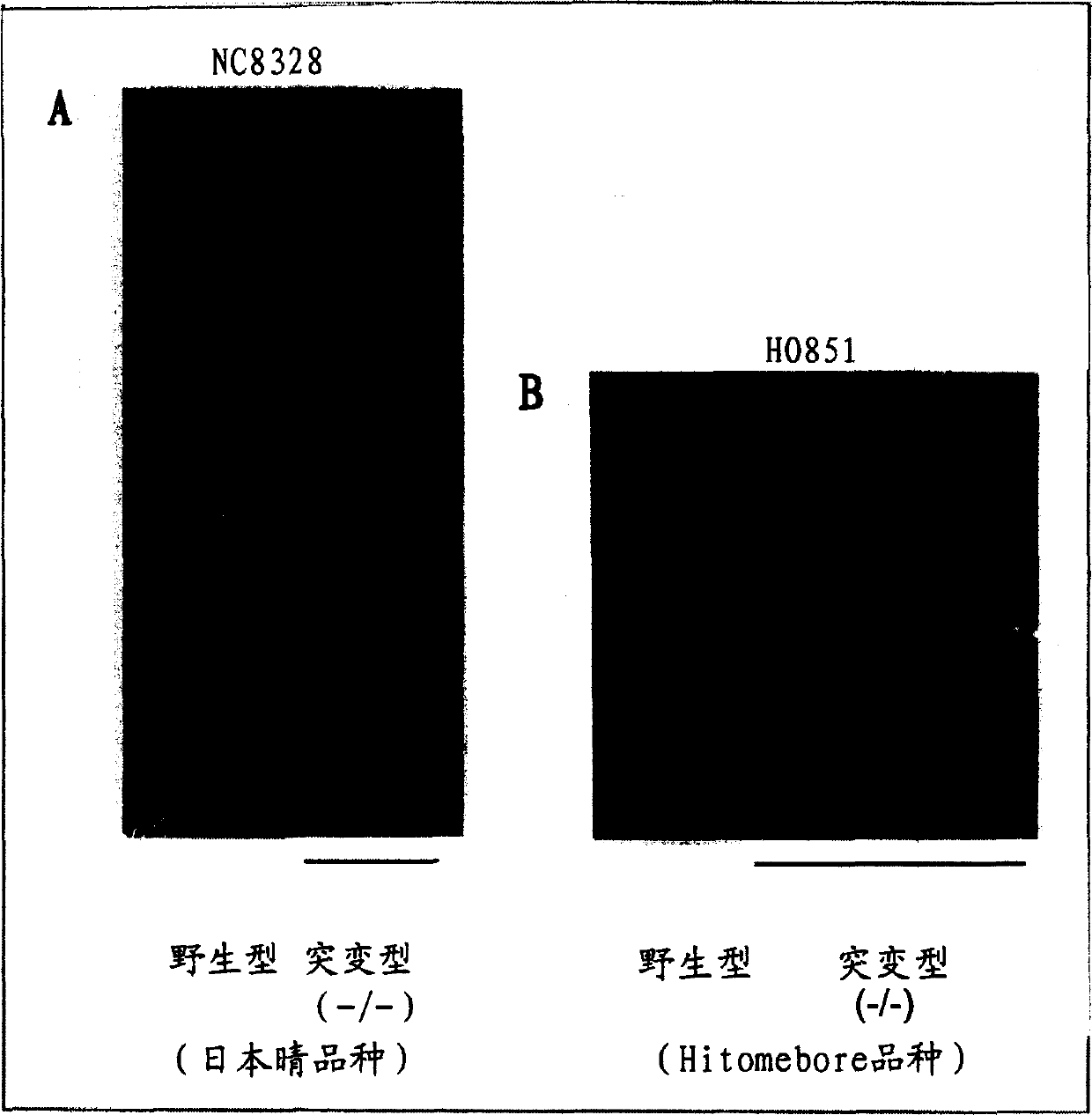
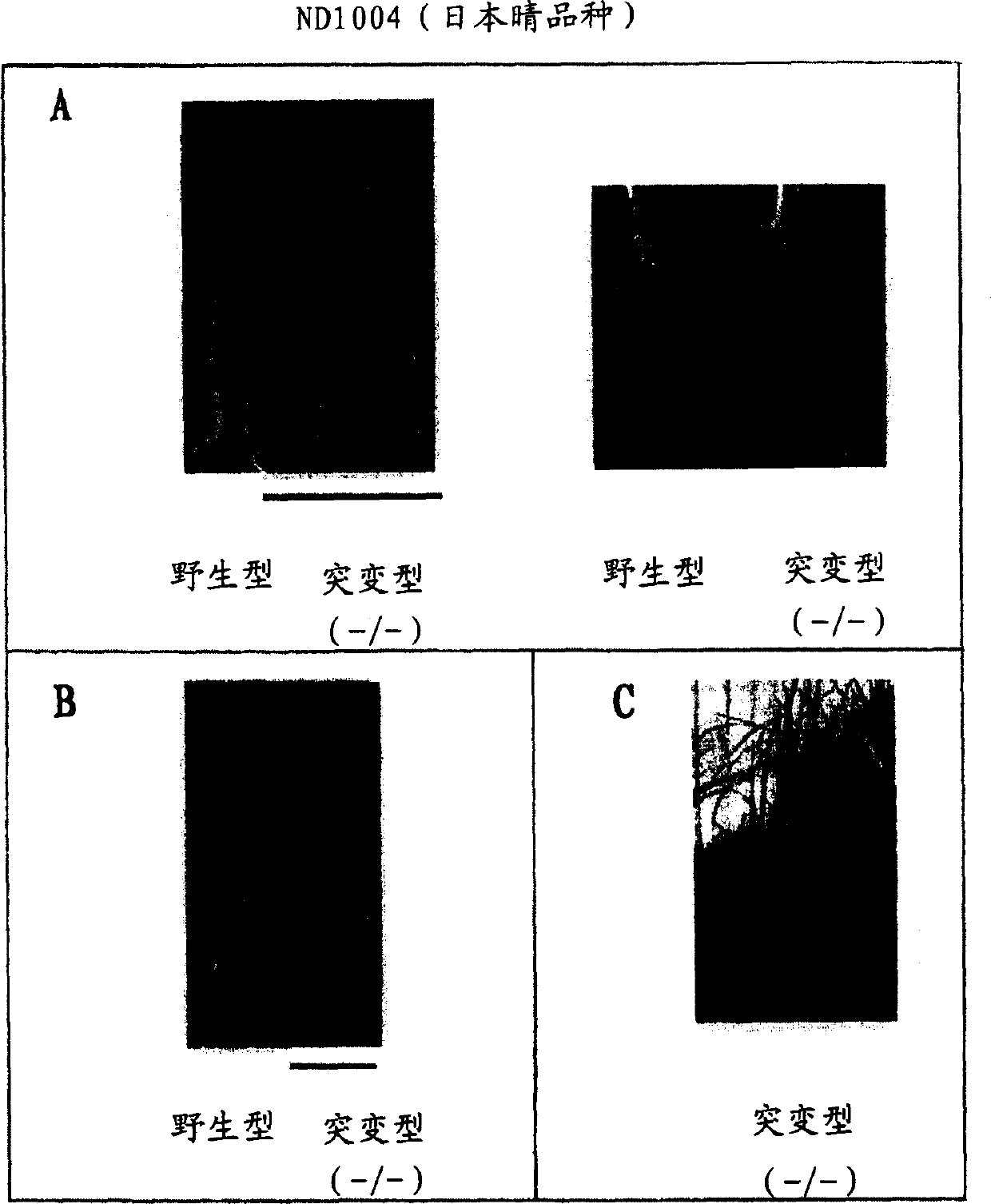
Novel rice gene regulating salt stress tolerance
A tolerant, salt-stressed technology, applied in a new field of genes, that can solve problems such as salt water influx, high financial or human cost, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0086] (Example 1: Activation of Tosl7 by culture and identification of the resulting mutants)
[0087] As described in Hirochika et al, 1996, Proc. For callus initiation culture and cell suspension culture. According to the method of Otsuki (1990) (rice protoplast culture line, Association for Agriculture, Forestry and Fisheries Technology Information), the culture conditions for activating Tosl7 for gene disruption were determined.
[0088] Briefly, mature rice seeds (Otsuki (1990), supra) were cultured in MS medium containing 2,4-dichlorophenoxyacetic acid (2,4-D) (25°C, 1 month) , to induce callus. The resulting callus was cultured in N6 liquid medium containing 2,4-D (Otsuki (1990), supra) for 5 months, and the callus was transferred to the redifferentiation medium (Otsuki (1990), Reference supra), to obtain redifferentiated rice (first generation (R1) plants).
[0089] About 20 R1 seeds were recovered from each plant. Seeds were sterilized with 1.0% sodium hypochlor...
Embodiment 2
[0091] (Example 2: Isolation of flanking sequences of Tosl7)
[0092] To find the genes controlling the phenotypes observed in Example 1, it was necessary to isolate the Tosl7 flanking sequences that had been transferred into the genomic DNA.
[0093] DNA was prepared from the R2 rice (line ND1004) obtained in Example 1 by the CTAB method (Murray and Thompson, 1980, Nucleic Acids Res, 8, 4321-4325). Tosl7 target site sequences were amplified by inverse PCR using total DNA as described (Hirochika et al., 1996, supra; and Sugimoto et al., 1994, Plant J., 5, 863-871).
[0094] Briefly, approximately 0.5 μg of total DNA from mutant plants (ND1004- / - lines) in which Tosl7 had been inserted into the target site by transposition was first digested with XbaI. Digested DNA was extracted with phenol / chloroform, followed by ethanol precipitation for purification. Ligation was then performed overnight using T4 DNA ligase in a total volume of 300 μl at 12°C. The ligated DNA was purified...
Embodiment 3
[0095] (Example 3: Structural analysis of the priming gene in the mutant)
[0096] RNA from wild-type rice (Nipponbare) seedlings grown in soil for 11 days was prepared as follows. First, total RNA of seedlings was extracted using ISOGEN solution. Total RNA was loaded onto an oligo(dt) cellulose column included in the mRNA purification kit (Stratagene) to obtain poly(A) mRNA. cDNA is synthesized from the obtained poly(A) mRNA by a usual method. The cDNA library was constructed in the HybriZAP-2.1 vector (Stratagene). The infectious capacity of the cDNA library was 5×10 5 a plaque. The pBluescript plasmid containing the cDNA insert was lysed in vivo using the E. coli strain XL1-Blue MRF2.
[0097]The cDNA library was screened according to the method described in Molecular Cloning, A Laboratory Manual (Sambrook et al., 1989), in which the inverse PCR product of Tosl7 flanking sequence obtained in Example 2 was used as a probe.
[0098] Nine cDNA clones showing strong hybri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com